Abstract
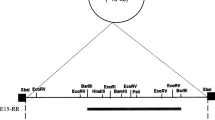
Rep22 is the replication region of the lactococcal theta replicating pUCL22 plasmid. The copy number of Rep22-based plasmids in Lactococcus was determined by using a chromosomal DNA fragment from Lactococcus lactis subsp. lactis MMS368 as reference. Segregational behavior appeared to be linked to copy number and therefore indicated random distribution of copies to daughter cells. Nevertheless, an active partitioning system was detected in the parental plasmid pUCL22. A pUCL22 138-bp DNA restriction fragment bearing a perfect 18-bp inverted repeat was involved in the improvement of Rep22-based plasmid segregational stability during discontinuous exponential growth.
Similar content being viewed by others
Literature Cited
Anderson DG, McKay LL (1983) Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol 46:549–552
Austin SJ, Eichorn BG (1992) Random diffusion can account for topA-dependent suppression of partition defects in lowcopy-number plasmids. J Bacteriol 174:5190–5195
Balke VL, Gralla JD (1987) Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol 169:4499–4506
Bremer H, Dennis PP (1987) Modulation of chemical composition and other parameters of the cell by growth rate. In: (Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE (eds) Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, DC: American Society for Microbiology, pp 1527–1542
D'angio C, Béal C, Boquien C-Y, Langella P (1994) Plasmid stability in recombinant strains of Lactococcus lactis subsp. lactis during continuous culture. FEMS Microbiol Lett 116: 25–30
El Alami N, Boquien C-Y, Corrieu G (1992) Batch cultures of recombinant Lactococcus lactis subsp. lactis in a stirred fermentor. Appl Microbiol Biotechnol 37:358–363
Feirtag JM, Petzel JP, Pasalados E, Baldwin KA, McKay LL (1991) Thermosensitive plasmid replication, temperaturesensitive host growth, and chromosomal plasmid integration conferred by Lactococcus lactis subsp. cremoris lactose plasmids in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 57:539–548
Frère J, Benachour A, Novel M (1993a) Identification of the theta-type minimal replicon of the Lactococcus lactis subsp. lactis CNRZ270 lactose protease plasmid pUCL22. Curr Microbiol 27:97–102
Frère J, Novel M, Novel G (1993b) Molecular analysis of the Lactococcus lactis subspecies lactis CNRZ270 bidirectional theta replicating lactose plasmid pUCL22. Mol Microbiol 10:1113–1124
Gasson MJ (1983) Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9
Kiewiet R, Kok J, Seegers JF, Venema G, Bron S (1993) The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl Environ Microbiol 59:358–354
Kuhl SA, Larsen LD, McKay LL (1979) Plasmid profiles of lactose-negative and proteinase-deficient mutants of S. lactis C10, ML3 and M18. Appl Environ Microbiol 37:1193–1195
Kunji ERS, Ubbink T, Matin A, Poolman B, Konings WN (1993) Physiological responses of Lactococcus lactis ML3 to alternating conditions of growth and starvation. Arch Microbiol 159:372–379
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor, N.Y., Cold Spring Harbor Laboratory Press
Sinha RP (1989) A new simple method of curing plasmids in lactic streptococci. FEMS Microbiol Lett 57:349–352
Terzaghi BE, Sandine WE (1975) Improved medium for lactic streptococci and their bacteriophages. Appl Environ Microbiol 29:807–813
von Wright A, Wessels S, Tynkkynen S, Saarela M (1990) Isolation of a replication region of a large lactococcal plasmid and use in cloning of a nisin resistance determinant. Appl Environ Microbiol 56:2029–2035
Weichart D, Lange R, Henneberg N, Hengge-Aronis R (1993) Identification and characterisation of stationary phaseinducible genes in Escherichia coli. Mol Microbiol 10:407–420
Williams DR, Thomas CM (1992) Active partitioning of bacterial plasmids. J Gen Microbiol 138:1–16
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Frère, J., Herreman, C., Boutibonnes, P. et al. Segregational stability and copy number of the theta-type lactococcal replicon Rep22 in Lactococcus . Current Microbiology 30, 33–37 (1995). https://doi.org/10.1007/BF00294521
Issue Date:
DOI: https://doi.org/10.1007/BF00294521