Abstract
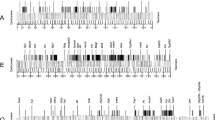
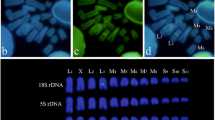
To study the middle repetitive fraction of the Drosophila subobscura genome, 26 phage clones containing repetitive sequences were examined by Southern DNA blot analysis and by in situ hybridization to polytene chromosomes. These results led to a classification of the clones according to five different types of hybridization patterns. Two types, each containing seven clones, are characterized by hybridization at 100 to 300 sites dispersed over the euchromatic parts of the chromosomes, and in addition by one prominently labelled chromosome band. One of these two classes also showed strong labelling of the chromocentre. The remaining types of hybridization pattern lacked a prominent band but showed hybridization either to the euchromatic regions or to the chromocentre or both. Chromosome A (=X) was the preferred location of prominently labelled bands and it also showed an excess of labelling by some clones. Some of the cloned dispersed sequences were localized cytologically on chromosomes of larvae from crosses between different strains of D. subobscura and between two closely related species, in order to detect heterozygosity at hybridization sites. Comparisons of the chromosomal distribution of labelling sites showed differences in number and location, indicating the possibility of transposition events.
Similar content being viewed by others
References
Anderson WW, Dobzhansky T, Pavlowsky O, Powell J, Yardley D (1975) Genetics of natural populations. XLII. Three decades of genetic changes in Drosophila pseudoobscura. Evolution 29:24–36
Anxolbéhère D, Nouaud D, Periquet G (1985) Séquences homologues à l'élément P chez des éspèces de Drosophila du groupe obscura et chez Scaptomyza pallida (Drosophilidae). Genet Sel Evol 17:579–584
Atherton D, Gall J (1972) Salivary gland squashes for in situ nucleic acid hybridization studies. Dros Inf Ser 49:131–133
Böhm I, Pinsker W, Sperlich D (1987) Cytogenetic mapping of marker genes on the chromosome elements C and E of Drosophila pseudoobscura and D. subobscura Getica 75:89–101
Dawid IB, Long EO, DiNocera PP, Pardue ML (1981) Ribosomal insertion-like elements in Drosophila melanogaster are interspersed with mobile elements. Cell 25:399–408
Dobzhansky Th (1970) Genetics and the evolutionary process. Columbia University Press, New York
Engels WR, Preston CR (1984) Formation of chromosome rearrangements by P factors in Drosophila. Genetics 107:657–678
Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Felger I, Pinsker W (1987) Histone gene transposition in the phylogeny of the Drosophila obscura group. Z Zool Syst Evolutions-forsch 25:127–140
Finnegan DJ (1985) Transposable elements in eukaryotes. Int Rev Cytol 93:281–326
Frischauf A, Lehrach H, Poustka A, Murray N (1983) Lambda replacement vectors carrying polylinker sequences. J Mol Biol 170:827–841
Huijser P, Hennig W, Dijkhof R (1987) Poly(dC-dA/dG-dT) repeats in the Drosophila genome: a key function for dosage compensation and position effect. Chromosoma 95:209–215
Krimbas CB, Loukas M (1980) The inversion polymorphism of Drosophila subobscura. Evol Biol 12:163–234
Krimbas CB, Loukas M (1984) Evolution of the obscura group Drosophila species. I. Salivary gland chromosomes and quantitative characters in D. subobscura and two closely related species. Heredity 53:469–482
Kunze-Mühl E, Müller E (1958) Weitere Untersuchungen über die chromosomale Struktur und die natürlichen Strukturtypen von Drosophila subobscura Coll. Chromosoma 9:559–570
Kunze-Mühl E, Sperlich D (1955) Inversionen und chromosomale Strukturtypen bei Drosophila subobscura. Z Indukt AbstammVererbungslehre 87:65–84
Lifschytz E, Hareven D (1982) Heterochromatic markers: A search for heterochromatin specific middle repetitive sequences in Drosophila. Chromosoma 86:429–442
Loukas M, Kafatos FC (1986) The actin loci in the genus Drosophila: Establishment of chromosomal homologies among distantly related species by in situ hybridization. Chromosoma 94:297–308
Loukas M, Kafatos FC (1988) Chromosomal locations of actin genes are conserved between the melanogaster and obscura groups of Drosophila. Genetica 76:33–41
Lowenhaupt K, Rich A, Pardue ML (1989) Nonrandom distribution of long mono and dinucleotide repeats in Drosophila chromosomes: Correlations with dosage compensation, heterochromatin and recombination. Mol Cell Biol 9:1173–1182
Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Manning JE, Schmid CW, Davidson N (1975) Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell 4:141–155
Martin G, Wiernasz D, Schedl P (1983) Evolution of Drosophila repetitive-dispersed DNA. J Mol Evol 19:203–213
Moltó MD, De Frutos R, Martinez-Sebastian MJ (1987) The banding pattern of polytene chromosomes of Drosophila guanche compared with that of D. subobscura. Genetica 75:55–70
Pardue ML (1986) In situ hybridization to DNA of chromosomes and nuclei. In: Roberts DB (ed) Drosophila — a practical approach. IRL press, Oxford, UK, pp 111–136
Pinsker W, Sperlich D (1979) Allozyme variation in natural populations of Drosophila subobscura along a north-south gradient. Genetica 50:207–219
Pinsker W, Sperlich D (1984) Cytogenetic mapping of enzyme loci on chromosomes J and U of Drosophila subobscura. Genetics 108:913–926
Preiss A, Hartley DA, Artavanis-Tsakonas S (1988) A molecular genetics of Enhancer of split, a gene required for embryonic neutral development in Drosophila. EMBO J 12:3917–3927
Rigby PWJ, Dieckmann M, Rhodes C, Berg P (1977) Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol 113:237–251
Roiha H, Miller JR, Woods LC, Glover DM (1981) Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature 290:749–753
Southern E (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 90:503–517
Sprading AC, Rubin GM (1981) Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet 15:219–264
Steinemann M, Pinsker W, Sperlich D (1984) Chromosome homologies within the Drosophila obscura group probed by in situ hybridization. Chromosoma 91:46–53
Syvanen M (1984) The evolutionary implications of mobile genetic elements. Annu Rev Genet 18:271–293
Traverse KL, Pardue ML (1989) Studies of He-T DNA sequences in the pericentric regions of Drosophila chromosomes. Chromosoma 97:261–271
Wensink PC, Tabata S, Pachl C (1979) The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell 18:1231–1246
Young MW (1979) Middle repetitive DNA: A fluid component of the Drosophila genome. Proc Natl Acad Sci USA 76:6274–6278
Young MW, Schwartz HE (1980) Nomadic gene families in Drosophila. Cold Spring Harbor Symp Quant Biol 45:629–640
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Felger, I., Sperlich, D. Cytological localization and organization of dispersed middle repetitive DNA sequences of Drosophila subobscura . Chromosoma 98, 342–350 (1989). https://doi.org/10.1007/BF00292387
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00292387