Summary
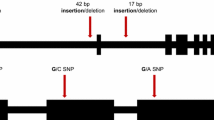
Speed, efficiency, and safety considerations have led many genome mapping projects to evaluate polymerase chain reaction (PCR) sequence amplification as an alternative to Southern blot analysis. However, the availability of informative primer sequences can be a limiting factor in PCR-based mapping. An alternative to random amplified polymorphism detection (RAPD) is the sequence-tagged-site (STS) approach. If informative primer sequences could be derived from known sequences, then current maps, which are based on both known function and anonymous clones, might be easily converted to maps utilizing PCR technology. In this paper, four pairs of primer sequences were obtained from published sequences, and four pairs were obtained by sequencing portions of DNA clones from genomic clones derived from a random genomic library used in the North American Barley Genome Mapping Project (NABGMP). These primers were used to screen for polymorphisms in the progeny of a winter x spring and a spring x spring barley cross. Two types of polymorphisms were distinguished using these primer sets: (1) insertion/deletion events that could be read directly from agarose gels, and (2) point mutation events. The latter were identified using polyacrylamide-gel electrophoresis of PCR products following digestion with restriction endonucleases (four-base cutters). To determine whether the PCR-based polymorphisms were allelic to polymorphisms identified by the clones from which the primer sequences derived, chromosomal assignments and (when possible) co-segregation analysis was performed.
Similar content being viewed by others
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1987) Current protocol in molecular biology. Greene Publishing Associates and Wiley-Interscience, pp 2.3.1–2.3.3
Beckmann JS (1988) Oligonucleotide polymorphisms: a new tool for genomic genetics. Bio/Technology 6:1061–1063
Beckmann JS, Soller M (1983) Restriction fragment length polymorphisms in genetic improvement: methodologies, mapping and costs. Theor Appl Genet 67:35–43
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Cole CG, Goodfellow PN, Bobrow M, Bentley DR (1991) Generation of novel sequence tagged sites (STSs from discrete chromosomal region using Alu-PCR. Genomics 10:816–826
D'Ovidio R, Tanzarella OA, Proceddu E (1990) Rapid and efficient detection of genetic polymorphisms in wheat through amplification by polymerase chain reaction. Plant Mol Biol 15:169–171
Forde BG, Heyworth A, Pywell J, Miflin BJ (1985) Nucleotide sequence of a B1-hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucleic Acids Res 13:7327–7339
Gyllensten UB, Erlich HA (1988) Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQalpha locus. Proc Natl Acad Sci USA 85:7652–7656
Higuchi R, Von Beroldingen CH, Sensabaugh GF, Erlich HA (1988) DNA typing from single hairs. Nature 332:543–546
Higuchi RG, Ochman H (1989) Production of single-stranded DNA templates by exonuclease digestion following the polymerase chain reaction. Nucleic Acids Res 17:5865
Innis MA, Myambo KB, Gelfand DH, Brow MAD (1988) DNA sequencing with Thermus aquatics DNA polymerase, and direct sequencing of PCR-amplified DNA. Proc Natl Acad Sci USA 85:9436–9440
Islam AKRM, Shepherd KW, Sparrow DHB (1981) Isolation and characterization of euplasmic wheat-barley chromosome addition lines. Heredity 46:161–174
Kurt JH, Bowcock AM, Erlich HA, Nevo S, Cavalli-Sforza LL (1991) Km typing with PCR: application to population screening. Am J Genet 48:613–620
Kusukawa NT, Uemori T, Asada K, Kato I (1990) Rapid and reliable protocol for direct sequencing of material amplified by PCR. Biotechniques 66:2–6
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Li H, Gyllensten UB, Cui X, Saiki RK, Erlich HA, Arnheim N (1988) Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335:414–417
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in-vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet 44:397–401
Mullis KB, Faloona F (1987) Specific synthesis of DNA in vitro via a polymerase chain reaction. Methods Enzymol 155:335–350
Myers RM, Maniatis T, Lerman LS (1987) Detection and localization of single-base changes by denaturing gradient-gel electrophoresis. Methods Enzymol 155:501–527
Olson M, Hood L, Cantor C, Doststein D (1989) A common language for physical mapping of the human genome. Science 254:1434–1435
Riedel GE, Swanberg SL, Kuranda KD, Marquette K, LaPan P, Bledsoe P, Kennedy A, Lin BY (1990) Denaturing gradient-gel electrophoresis identifies genomic DNA polymorphism with high frequency in maize. Theor Appl Genet 80:1–10
Rodriguez-Palnezuela P, Pintor-Toro J, Carbonero P, Garcian Olmedo F (1988) Nucleotide sequence and endosperm-specific expression of the structural gene for the toxin α-hordothionin in barley (Hordeum vulgare L). Gene 70:271–281
Saiki RK, Bugawan GT, Horn KB, Mullis KB, Erlich HA (1986) Analysis of enzymatically amphified β-globin and HLA-DQ DNA with allele-specific oligonucleotide probes. Nature 324:163–164
Saiki RK, Scarf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle-cell anemia. Science 230:1350–1354
Sanger FS, Nicklen S, Coulson AR (1977) DNA sequencing with chaintermination inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Shin JS, Chao S, Corpuz L, Blake TK (1990) A partial map of the barley genome incorporating restriction fragment length polymorphism, polymerase chain reaction, isozyme, and morphological marker loci. Genome 33:803–810
Skolnick MH, Wallace RB (1988) Simultaneous analysis of multiple polymorphic loci using amplified sequence polymorphism (ASPs). Genomics 2:273–279
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Trick M, Dennis ES, Edwards KJK, Peacock WJ (1988) Molecular analysis of the alcohol dehydrogenase gene family of barley. Plant Mol Biol 11:147–160
Weber JL, May PE (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396
Weining S, Langridge P (1991) Identification and mapping of polymorphisms in cereal based on the polymerase chain reaction. Theor Appl Genet 82:209–216
Williams JGK, Kubelik ARK, Livak JL, Rafalsla JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Williams MNV, Pande N, Nair S, Mohan M, Bennett J (1991) Restriction fragment length polymorphism analysis of polymerase chain reaction products amplified from mapped loci of rice (Oryza sativa L.) genomic DNA. Theor Appl Genet 82:489–498
Wong C, Dowling CE, Saiki RK, Higuchi RG, Erlich HA, Kazazian HH (1987) Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single-copy DNA. Nature 330:384–386
Author information
Authors and Affiliations
Additional information
Communicated by J. W. Snape
Rights and permissions
About this article
Cite this article
Tragoonrung, S., Kanazin, V., Hayes, P.M. et al. Sequence-tagged-site-facilitated PCR for barley genome mapping. Theoret. Appl. Genetics 84, 1002–1008 (1992). https://doi.org/10.1007/BF00227417
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00227417