Summary
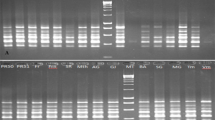
Restriction fragment length polymorphisms (RFLP) were assessed among accessions within six peanut species of the Arachis section: tetraploid cultivated species, A. hypogaea; tetraploid wild species, A. monticola; and four diploid wild species, A. batizocoi,A. cardenasii, A. duranensis and A. glandulifera. While the two tetraploid species did not show polymorphism with 16 PstI-generated random genomic probes, two of seven seed cDNA probes detected polymorphisms. The RFLP variation detected by two seed cDNA probes appeared to be related to structural changes occurring within tetraploid species. The botanical var. ‘fastigiata’ (Valencia market type) of A. hypogaea subspecies fastigiata was shown to be the most variable. Arachis monticola was found to be more closely related to A. hypogaea subspecies hypogaea than to subspecies fastigiata. Diploid species A. cardenasii, A. duranensis, and A. glandulifera showed considerable intraspecific genetic diversity, but A. batizocoi showed little polymorphism. The genetic distance between the cultivated peanut and wild diploid species was found to be closest for A. duranensis.
Similar content being viewed by others
References
Bernatzky R, Tanksley SD (1986) Genetics of actin related sequences in tomato. Theor Appl Genet 72:314–321
Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 88:1991–1995
Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Gregory WC, Gregory MP, Krapovickas A, Smith BW, Yarbrough JA (1973) Structures and genetic resources of peanuts. In: Summerfield RJ, Bunting AH (eds) Peanutsculture & uses. Am Peanut Res Educ Assoc, Stillwater Okla., pp 47–133
Hammons RO (1982) Origin and early history of the peanut. In: Pattee HE, Young CT (eds) Peanut science and technology. Am Peanut Res Educ Soc, Yoakum Tex., pp 1–20
Helentjaris T, King G, Slocum M, Siedenstrang C, Wegman S (1985) Restriction fragment polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol Biol 5:109–118
Herman J, Lee P, Saya H, Nakajima M (1990) Application of polymerase chain reaction for rapid subcloning of cDNA inserts from lambda gt11 clones. Biotechniques 8:376–379
Higgins TJV (1984) Synthesis and regulation of major proteins in seeds. Annu Rev Plant Physiol 35:191–221
Husted L (1936) Cytological studies of peanut, Arachis. II. Chromosome number, morphology and behavior and their application to the origin of cultivated forms. Cytologia 7:396–423
Ish-Horowicz D, Burke JF (1981) Rapid and efficient cosmid cloning. Nucleic Acids Res 9: 2989
Keim P, Shoemaker RC, Palmer RG (1989) Restriction fragment length polymorphism diversity in soybean. Theor Appl Genet 77:786–792
Kochert G, Halward T, Branch WD, Simpson CE (1991) RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor Appl Genet 81:565–570
Nei M (1987) Genomic evolution, genetic variation within population, genetic variation between population. In: Molecular evolutionary genetics. Columbia University Press, New York, pp 111–253
Ohno S (1970) Why gene duplication, Mechanisms of gene duplication. In: Evolution by gene duplication. Springer, Berlin Heidelberg New York, pp 59–110
Riley R, Chapman V, Kimber G (1960) Position of the gene determining the diploid-like meiotic behavior of wheat. Nature 186:259–260
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014
Schaffer HE, Sederoff RR (1981) Improved estimation of DNA fragment lengths from agarose gel. Anal Biochem 115:113–122
Seetharam A, Nayar KMD, Sreekantaradhya R, Achar DKT (1973) Cytological studies on the interspecific hybrid of Arachis hypogaea x Arachis duranensis. Cytologia 38:277–280
Sharp PJ, Chao S, Desai S, Kilian A, Gale MD (1989) Use of RFLP markers in wheat and related species. In: Helentjaris T, Burr B (eds) Current communications in molecular biology-development and application of molecular markers to problems in plant genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y, pp 29–33
Singh AK (1986) Utilization of wild relatives in the genetic improvement of Arachis hypogaea L. 8. Synthetic amphidiploids and their importance in interspecific breeding. Theor Appl Genet 72:433–439
Singh AK, Moss JP (1982) Utilization of wild relatives in the genetic improvement of Arachis hypogaea L. Part 2. Chromosome complements of species in section Arachis. Theor Appl Genet 61:305–314
Singh AK, Moss JP (1984) Utilization of wild relatives in the genetic improvement of Arachis hypogaea L. 5. Genome analysis in section Arachis and its implications in gene transfer. Theor Appl Genet 68:335–364
Smartt J, Gregory WC, Gregory MP (1978) The genomes of Arachis hypogaea. 1. Cytogenetic studies of putative genome donors. Euphytica 27:665–675
Sneath PHA, Sokal RR (1973) Sequential, agglomerative, hierarchic, nonoverlapping clustering methods. In: Kennedy D, Park RB, Numerical taxonomy. W.H. Freeman, San Francisco, pp 201–240
Stalker HT, Moss JP (1987) Speciation, cytogenetics and utilization of Arachis species. Adv Agron 41:1–40
Stalker HT, Wynne JC (1979) Cytology of interspecific hybrids in section Arachis of peanut. Peanut Sci 6:110–114
Swofford DL, Selander RB (1981) Biosys-1; A computer program for the analysis of allelic variation in genetics. Department of Genetics and Development, University of Illinois at Urbana-Champaign, Ill.
Author information
Authors and Affiliations
Additional information
Communicated by P. L. Pfahler
Florida Agricultural Experiment Station, Journal Series No. R-01493
Rights and permissions
About this article
Cite this article
Paik-Ro, O.G., Smith, R.L. & Knauft, D.A. Restriction fragment length polymorphism evaluation of six peanut species within the Avachis section. Theoret. Appl. Genetics 84, 201–208 (1992). https://doi.org/10.1007/BF00224001
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00224001