Abstract
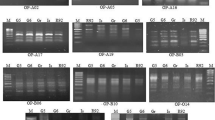
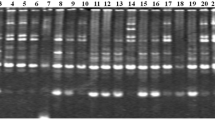
Ribes nigrum germplasm was screened for random amplified polymorphic DNA (RAPD) markers. Fiftyfour markers were identified which generated individual fingerprints for each of 21 cultivars. Genetic variation within R. nigrum germplasm, as detected by RAPDs, demonstrated that the genetic basis for improvement of blackcurrant is narrower than would be expected by the analysis of parentage.
Similar content being viewed by others
References
Barua UM, Chalmers KJ, Hackett CA, Thomas WTB, Powell W, Waugh R (1993) Identification of RAPD markers linked to a Rhynchosporium secalis resistance locus in barley using nearisogenic lines and bulked segregant analysis. Heredity 71:177–184
Collins GG, Symons RH (1993) Polymorphism in grapevine detected by the RAPD PCR technique. Plant Mol Biol Rep 11:105–112
Demeke T, Adams RP, Chibbar R (1992) Potential taxonomic use of random amplified polymorphic DNA (RAPD): a case study in Brassica. Theor Appl Genet 84:990–994
Eskew DL, Caetano-Anolles G, Bassam BJ, Gresshoff PM (1993) DNA amplification fingerprinting of the Azolla-Anabaena symbiosis. Plant Mol Biol 21:363–373
Gawel NJ, Jarret RL (1991) A modified CTAB DNA extraction procedure for Musa and Ipomoea. Plant Mol Biol Rep 9:262–266
Graham J, McNicol RJ, Greig K, Van de Ven WTG (1994) Identification of red raspberry cultivars and assessment of their relatedness using fingerprints produced by random primers. J Hortic Sci 69:123–130
Halward T, Stalker T, LaRue E, Kochert G (1992) Use of singleprimer DNA amplifications in genetic studies of peanut (Arachis hypogaea L.). Plant Mol Biol 18:315–325
Harada T, Matsukawa K, Sato T, Ishikawa R, Niizeki M, Saito K (1993) DNA-RAPDs detect genetic variation and paternity in Malus. Euphytica 65:87–91
Kazan K, Manners JM, Cameron DF (1993) Genetic relationships in the Stylosanthes guianensis species complex assessed by random amplified polymorphic DNA. Genome 36:43–49
Kresovich S, Williams JGK, McFerson JR, Routman EJ, Schaal BA (1992) Characterization of genetic identities and relationships of Brassica oleracea L. via a random amplified polymorphic DNA assay. Theor Appl Genet 85:190–196
Lihonos FD, Pavlova NM (1969) Fruit crops (in Russian). Tr Prikl Bot Genet Sel 41:264–284
Malecot G (1948) Les mathematiques de l'heredite. Massom and Cie, Paris
Martin GB, Williams JGK, Tanksley SD (1991) Rapid identification of markers linked to a Pseudomonas resistance gene in tomato by using random primers and near-isogenic lines. Proc Natl Acad Sci USA 88:2336–2340
Melekhina AA (1964) Varieties and the breeding of blackcurrants (in Russian). Sadovodstvo 12:33
Mosolova AV (1963) Siberian plant breeders in Leningrad province (in Russian). Sadovodstvo 3:33–35.Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76: 5267–5273
Paran I, Michelmore RW (1993) Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85:985–993
Pavlova NM, Volodina EV (1978) Features of morphological characters in blackcurrant varieties (in Russian). Tr Prikl Bot Genet Sel 62:102–109
Powell W, Phillips MS, McNicol JW, Waugh R (1991) The use of DNA markers to estimate the extent and nature of genetic variability in Solanum tuberosum cultivars. Ann Appl Biol 118:423–432
Quiros CF, Hu J, This P, Chevre AM, Delseny M (1991) Development and chromosomal localization of genome-specific markers by polymerase chain reaction in Brassica. Theor Appl Genet 82:627–632
Reiter RS, Williams JGK, Feldmann KA, Rafalski JA, Tingey SD, Scolnik PA (1992) Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lined and random amplified polymorphic DNAs. Proc Natl Acad Sci USA 89:1477–1481
Roy A, Frascaria N, MacKay J, Bousquet J (1992) Segregating random amplified polymorphic DNAs (RAPDs) in Betula alleghaniensis. Theor Appl Genet 85:173–180
Scott M, Haymes M, Williams SM (1992) Parentage analysis using RAPD PCR. Nucleic Acids Res 20:5493
Smith CE (1969) From Vavilov to the present — a review. Econ Bot 23:2–19.
Tydeman HM (1938) Some results of experiments in breeding blackcurrants. 2. First crosses between the main varieties. J Pomol 16:224–250.
Vitkovskij VL (1964a) Small fruit breeding under conditions of the Far North (in Russian). Tr Prikl Bot Genet Sel 36:149–157
Volunez AG (1988) Breeding interesting cultivars of blackcurrant (in Russian). Sadovodstvo 8:28
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acid Res 18:7213–7218
Welsh J, Hoercutt RJ, McClellend M, Sobral BWS (1991) Parentage determination in maize hybrids using the arbitrarily primed polymerase chain reaction (AP-PCR). Theor Appl Genet 82:473–476
Wilde J, Waugh R, Powell W (1992) Genetic fingerprinting of Theobroma clones using randomly amplified polymorphic DNA markers. Theor Appl Genet 83:871–877
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Williams JGK, Reiter RS, Young RM, Scolnik PA (1993) Genetic mapping of mutations using phenotypic pools and mapped RAPD markers. Nucleic Acids Res 21:2697–2702
Yang X, Quiros C (1993) Identification and classification of celery cultivars with RAPD markers. Theor Appl Genet 86:205–212
Yu K, Pauls KP (1993) Rapid estimation of genetic relatedness among heterogeneous populations of alfalfa by random amplification of bulked genomic DNA samples. Theor Appl Genet 86:788–794
Zagaja SW (1983) Germplasm resources and exploration. In: Moore JN, Janick J (eds) Methods in fruit breeding. Indiana, Purdue University Press, pp 3–10
Author information
Authors and Affiliations
Additional information
Communicated by P. M. A. Tigerstedt
Rights and permissions
About this article
Cite this article
Lanham, P.G., Brennan, R.M., Hackett, C. et al. RAPD fingerprinting of blackcurrant (Ribes nigrum L.) cultivars. Theoret. Appl. Genetics 90, 166–172 (1995). https://doi.org/10.1007/BF00222198
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00222198