Abstract
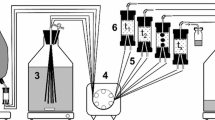
In this segment of a larger multidisciplinary study of the movement and fate of creosote derived compounds in a sand-and-gravel aquifer, we present evidence that the methanogenic degradation of the major biodegradable phenolic compounds and concomitant microbial growth in batch microcosms derived from contaminated aquifer material can be described using Monod kinetics. Substrate depletion and bacterial growth curves were fitted to the Monod equations using nonlinear regression analysis. The method of Marquardt was used for the determination of parameter values that best fit the experimental data by minimizing the residual sum of squares. The Monod kinetic constants (μ max , K s, Y, and k d) that describe phenol, 2-, 3-, and 4-methylphenol degradation and concomitant microbial growth were determined under conditions that were substantially different from those previously reported for microcosms cultured from sewage sludge. The K s values obtained in this study are approximately two orders of magnitude lower than values obtained for the anaerobic degradation of phenol in digesting sewage sludge, indicating that the aquifer microorganisms have developed enzyme systems that are adapted to low nutrient conditions. The values for k d are much less than μ max, and can be neglected in the microcosms. The extremely low Y values, approximately 3 orders of magnitude lower than for the sewage sludge derived cultures, and the very low numbers of microorganisms in the aquifer derived microcosms suggest that these organisms use some unique strategies to survive in the subsurface environment.
Similar content being viewed by others
Abbreviations
- GC:
-
gas chromatography
- HPLC:
-
high performance liquid chromatography
- LBSSB:
-
likelihood based sum of squares boundaries
- MPN:
-
most probable number
- NLR:
-
nonlinear regression analysis
- OFAG:
-
oxygen free Argon gas
- PCP:
-
pentachlorophenol
- RSS:
-
residual sum of squares
- SRB:
-
sulfate reducing bacteria
References
Bard Y (1974) Nonlinear parameter estimation. Academic Press, New York
Bates DM & Watts DG (1988) Nonlinear regression analysis and its applications. Wiley, New York
Brock TD & O'Dea K (1977) Amorphous ferrous sulfide as a reducing agent for culture of anaerobes. Appl. Environ. Microbiol. 33: 254–256
Constantinides A (1987) Applied numerical methods with personal computers. McGraw-Hill, New York
Dills SS, Apperson A, Schmidt MR & Saier MH Jr (1980) Carbohydrate transport in bacteria. Microbiol. Rev. 44: 385–418
Ehrlich GG, Godsy EM, Goerlitz DF & Hult MF (1983) Microbial ecology of a creosote-contaminated aquifer at St Louis Park, Minnesota. Dev. Ind. Microbiol. 24: 235–245
Fedorak PM & Hrudey SE (1986) Anaerobic treatment of phenolic coal conversion wastewater in semicontinuous cultures. Water Res. 20: 113–122
Franks BJ (1988) Hydrogeology and flow of water in a sand and gravel aquifer contaminated by wood-preserving compounds, Pensacola, Florida. U.S. Geological Survey WRI Report 87-4260
Furutani A, Rudd JWM & Kelly CA (1984) A method for measuring the response of sediment microbial communities to environmental perturbations. Can. J. Microbiol. 30: 1408–1414
Gälli R (1987) Biodegradation of dichloromethane in waste water using a fluidized bed bioreactor. Appl. Microbiol. Biotechnol. 27: 206–213
Godsy EM (1980) Isolation of Methanobacterium bryantii from a deep aquifer by using a novel broth-antibiotic disk method. Appl. Environ. Microbiol. 39: 1074–1075
Godsy EM, Goerlitz DF & Grbić-Galić D (1992) Methanogenic biodegradation of creosote contaminants in natural and simulated ground water ecosystems. Ground Water 30: 232–242
Healy JB & Young LY (1978) Catechol and phenol degradation by a methanogenic population of bacteria. Appl. Environ. Microbiol. 35: 216–218
Healy JB, Young LY & Reinhard M (1980) Methanogenic decomposition of ferulic acid, a model lignin derivative. Appl. Environ. Microbiol. 39: 438–444
Kindred JS & Celia MA (1989) Contaminant transport and biodegradation 2. Conceptual model and test simulations. Water Res. Research 25: 1149–1159
Monod J (1949) The growth of bacterial cultures. Ann. Rev. Microbiol. 3: 371–394
Mackey JK (1987) The influence of microbial dynamics on the steady state biodegradation of 2-chlorophenol in continuous culture. MENGR Report, College of Engineering, Clemson University, Clemson, S.C.
Neufeld RD, Mack JD & Strakey JP (1980) Anaerobic phenol biokinetics. J.Water Poll. Control Fed. 52: 2367–2377
Roberts DJ, Fedorak PM & Hrudey SE (1986) Comparison of the fates of the methyl carbons of m-cresol and p-cresol in methanogenic consortia. Can. J. Microbiol. 33: 335–338
Robinson JA (1985) Nonlinear regression analysis in microbial ecology. Adv. Microbial Ecol. 8: 61–114
Suidan MT, Najm IN, Pfeffer JT & Wang YT (1989) Anaerobic biodegradation of phenol: inhibition kinetics and system stability. J. Environ. Engng. 114: 1359–1376
Tarvin D & Buswell AM (1934) The methane fermentation of organic acids and carbohydrates. J. Am. Chem. Soc. 56: 1751–1755
Templeton LL & Grady CPL Jr. (1988) Effects of culture history on the determination of biodegradation kinetics by batch and fed-batch techniques. J. Water Poll. Control Fed. 60: 651–658
Wolin EA, Wolin MJ & Wolfe RS (1963) Formation of methane by bacterial extracts. J. Biol. Chem. 238: 2882–2886
Young LY & Rivera MD (1985) Methanogenic degradation of four phenolic compounds. Water Res. 19: 1325–1332
Zeikus JG (1977) The biology of methanogenic bacteria. Bacteriol. Rev. 41: 514–541
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Godsy, E.M., Goerlitz, D.F. & Grbić-Galić, D. Methanogenic degradation kinetics of phenolic compounds in aquifer-derived microcosms. Biodegradation 2, 211–221 (1991). https://doi.org/10.1007/BF00114553
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00114553