Abstract
The gene encoding Arabidopsis thaliana aspartate kinase (ATP:L-aspartate 4-phosphotransferase, EC 2.7.2.4) was isolated from genomic DNA libraries using the carrot ak-hsdh gene as the hybridizing probe. Two genomic libraries from different A. thaliana races were screened independently with the ak probe and the hsdh probe. Nucleotide sequences of the A. thaliana overlapping clones were determined and encompassed 2 kb upstream of the coding region and 300 bp downstream. The corresponding cDNA was isolated from a cDNA library made from poly(A)+-mRNA extracted from cell suspension cultures. Sequence comparison between the Arabidopsis gene product and an AK-HSDH bifunctional enzyme from carrot and from the Escherichia coli thrA and metL genes shows 80%, 37.5% and 31.4% amino acid sequence identity, respectively. The A. thaliana ak-hsdh gene is proposed to be the plant thrA homologue coding for the AK isozyme feedback inhibited by threonine. The gene is present in A. thaliana in single copy and functional as evidenced by hybridization analyses.
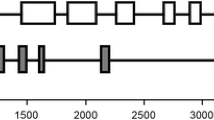
The apoprotein-coding region is interrupted by 15 introns ranging from 78 to 134 bp. An upstream chloroplast-targeting sequence with low sequence similarity with the carrot transit peptide was identified. A signal sequence is proposed starting from a functional ATG initiation codon to the first exon of the apoprotein. Two additional introns were identified: one in the 5′ non-coding leader sequence and the other in the putative chloroplast targeting sequence. 5′ sequence analysis revealed the presence of several possible promoter elements as well as conserved regulatory motifs. Among these, an Opaque2 and a yeast GCN4-like recognition element might be relevant for such a gene coding for an enzyme limiting the carbon-flux entry to the biosynthesis of several essential amino acids. 3′ sequence analysis showed the occurrence of two polyadenylation signals upstream of the polyadenylation site.
This work is the first report of the molecular cloning of a plant ak-hsdh genomic sequence. It describes a promoter element that may bring new insights to the regulation of the biosynthesis of the aspartate family of amino acids.
Similar content being viewed by others
Abbreviations
- AK:
-
aspartate kinase
- HSDH:
-
homoserine dehydrogenase
- ID:
-
intermediate domain
- Tp:
-
transit peptide
References
Cohen NG, Saint-Giron I: Biosynthesis of threonine, lysine and methionine. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE (eds) Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, vol I, pp. 429–444. American Society for Microbiology, Washington DC (1987).
Bryan JK: Advances in the biochemistry of amino acids. In: Miflin BJ, Lea PJ (eds) Intermediary Nitrogen Metabolism, pp. 161–195. The Biochemistry of Plants, vol. 16. Academic Press, New York (1990).
Hegel J, Frydrych Z: Optimum amino acid supplementation of a high-protein wheat. Lysine and threonine. Nutr Rep Int 36: 927–934 (1987).
Jacobs M, Frankard V, Ghislain M: Mutants in the biosynthesis of amino acids. In: Sangwan RS, Sangwan-Norreel BS (eds) The Impact of Biotechnology in Agriculture, pp. 247–258. Kluwer Academic Publishers, Dordrecht (1990).
Bright SWJ, Kueh JSH, Franklin J, Rognes SE, Miflin BJ: Two genes for threonine accumulation in barley seeds. Nature 299: 278–279 (1982).
Cattoir-Reynaerts A, Degryse E, Verbruggen I, Jacobs M: Selection and characterization of carrot embryoid cultures resistant to inhibition by lysine plus threonine. Biochem Physiol Pflanzen 178: 81–90 (1983).
Diedrick TJ, Frisch DA, Gengenbach BG: Tissue culture isolation of a second mutant locus for increased threonine accumulation in maize. Theor Appl Genet 79: 209–215 (1990).
Frankard V, Ghislain M, Negrutiu I, Jacobs M: High threonine producer mutant of Nicotiana sylvestris (Spegazzini and Comes). Theor Appl Genet 82: 273–282 (1991).
Vernaillen S, Van Ghelue M, Verbruggen I, Jacobs M: Characterization of mutants in Arabidopsis thaliana (L. Heyn.) resistant to analogues and inhibitory concentrations of amino acids derived from aspartate. Arab Inf Serv 22: 13–22 (1985).
Bright SWJ, Miflin BJ, Rognes SE: Threonine accumulation in the seeds of a barley mutant with an altered aspartate kinase. Biochem Genet 20: 229–243 (1982).
Relton JM, Bonner PLR, Wallsgrove RM, Lea PJ: Physical and kinetic properties of lysine-sensitive aspartate kinase purified from carrot cell suspension culture. Biochim Biophys Acta 953: 48–60 (1988).
Dotson SB, Somers DA, Gengenbach BG: Purification and characterization of lysine-sensitive aspartate kinase from maize cell cultures. Plant Physiol 91: 1602–1608 (1989).
Wilson BJ, Gray AC, Matthews BJ: Bifunctional protein in carrot contains both aspartokinase and homoserine dehydrogenase activities. Plant Physiol 97: 1323–1328 (1991).
Weisemann JM, Matthews BJ: Identification and expression of a cDNA from Daucus carota encoding a bifunctional aspartokinase-homoserine dehydrogenase. Plant Mol Biol 22: 301–312 (1993).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual; 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Hanahan D: Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557 (1983).
Dellaporta SL, Wood J, Hicks JB: A plant DNA mini preparation: version II. Plant Mol Biol Rep 1: 19–21 (1983).
Fourney RM, MiyaKoshi J, DayIII RS, Paterson MC: Northern blotting: efficient RNA staining and transfer. Focus 10: 5–7 (1987).
Sanger FS, Nicklen S, Coulson AR: DNA sequencing with the chain terminating inhibitors. Proc Natl Acad USA 74: 5463–5467 (1977).
Bilang R, Schnorf M: Mesophyll protoplasts of tobacco. In: Potrykus I (ed) Gene Transfer to Plants, pp. 18–25. EMBO Advanced Laboratory Course, Swiss Federal Institut of Technology (1991).
Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Keegstra K, Olsen LJ, Theg SM: Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Plant Physiol Plant Mol Biol 40: 471–501 (1989).
Brown JWS: A catalogue of splice junction and putative branch point sequences from plant introns. Nucl Acids Res 14: 9549–9559 (1986).
Kaneko T, Hashimoto T, Kumpaisal R, Yamada Y: Molecular cloning of wheat dihydrodipicolinate synthase. J Biol Chem 265: 17451–17455 (1990).
Cossart P, Katinka M, Yaniv M, Saint-Girons I, Cohen GN: Nucleotide sequence of the thr B gene of E. coli, and its two adjacent regions: the thr AB and thr C junctions. Nucl Acids Res 9: 339–347 (1981).
Zakin MM, Duchange N, Ferrara P, Cohen GN: Nucleotide sequence of the met L gene of Escherichia coli. Its product, the bifunctional aspartokinase II-homoserine dehydrogenase II and the bifunctional product of the thr A gene, aspartokinase I-homoserine dehydrogenase I derive from a common ancestor. J Biol Chem 258: 3028–3031 (1983).
Cassan M, Parsot C, Cohen GN, Patte JC: Nucleotide sequence of the lysC gene encoding the lysine sensitive aspartokinase III of Escherichia coli K12: evolutionary pathway leading to three isofunctional enzymes. J Biol Chem 261: 1052–1057 (1986).
Omori K, Imai Y, Suzuki S-I, Komatsubara S: Nucleotide sequence of the Serratia marcescens threonine operon and analysis of the threonine operon mutations which alter feedback inhibition of both aspartokinase I and homoserine dehydrogenase I. J Bact 175: 785–794 (1993).
Chen NY, Hu FM, Paulus H: Nucleotide sequence of the overlapping genes for the subunits of Bacillus subtilis aspartokinase II and their control regions. J Biol Chem 262: 8787–8798 (1987).
Kalinowski J, Bachmann B, Thierbach G, Pühler A: Aspartokinase genes lysCa and lysCb overlap and are adjacent to the aspartate β-semialdehyde dehydrogenase gene asd in Corynebacterium glutamicum. Mol Gen Genet 224: 317–324 (1990).
Kalinowski J, Cremer J, Bachmann B, Eggeling L, Sahm H, Pühler A: Genetic and biochemical analysis of the aspartokinase from Corynebacterium glutamicum. Mol Microbiol 5: 1197–1204 (1991).
Rafalski AJ, Falco CS: Structure of the yeast hom3 gene which encodes for aspartokinase. J Biol Chem 263: 2146–2151 (1988).
Hanley BA, Schuler MA: Plant intron sequences: evidence for distinct groups of introns. Nucl Acids Res 16: 7159–7175 (1988).
Omori K, Suzuki S-I, Imai Y, Komatsubara S: Analysis of the proBA operon and feedback control of proline biosynthesis. J Gen Microbiol 137: 509–517 (1991).
Deutch AH, Rushlow KE, Smith CJ: Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucl Acids Res 12: 6337–6355 (1984).
Hu C-A, Delauney AJ, Verma DPS: A bifunctional enzyme (D1-pyrroline-5-carboxylate synthetase) catalyses the first two steps in proline biosynthesis in plants. Proc Natl Acad Sci USA 89: 9354–9358 (1992).
Campbell WH, Gowri G: Codon usage in higher plants, green algae and cyanobacteria. Plant Physiol 92: 1–11 (1990).
Fazel A, Müller K, Le Bras G, Garel J-R, Veron M, Cohen GN: A triglobular model for the polypeptide chain of aspartokinase I-homoserine dehydrogenase I of Escherichia coli. Biochemistry 22: 158–165 (1983).
Joshi CP: An inspection of the domain between putative TATA box and translational start site in 79 plant genes. Nucl Acids Res 15: 6643–6653 (1987a).
Bucher P: Wheight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol 212: 563–578 (1990).
Arndt K, Fink GR: GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc Natl Acad Sci USA 83: 8516–8520 (1986).
Katagiri K, Chua N-H: Plant transcription factors: present knowledge and future challenges. Trends Genet 8: 22–27 (1992).
Forde BG, Heyworth A, Pywell J, Kreis M: Nucleotide sequence of a Bl hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucl Acids Res 13: 7327–7339 (1985).
Lohmer S, Maddaloni M, Motto M, Di Fonzo N, Hartings H, Salamini F, Thompson RD: The maize regulatory locus Opaque-2 encodes a DNA-binding protein which activates the transcription of the b-32 gene. EMBO J 10: 617–624 (1991).
Kodrzychi R, Boston RS, Larkins BA: The opaque 2 mutation of maize differentially reduces zein gene transcription. Plant Cell 1: 105–114 (1989).
Motto M, Di Fonzo N, Hartings H, Maddaloni M, Salamini F, Soave C, Thompson RD. Regulatory genes affecting maize storage protein synthesis. Oxford Surv Plant Mol Cell Biol 6: 87–114 (1989).
Bass HW, Webster C, OBrian GR, Roberts JKM, Boston RS: A maize ribosome-inactivating protein is controlled by the transcriptional activator Opaque 2. Plant Cell 4: 225–234 (1992).
Joshi CP: Putative polyadenylation signals in nuclear genes of higher plants: a compilation analysis. Nucl Acids Res 15: 9627–9640 (1987b).
Mogen BD, MacDonald MH, Graybosh R, Hunt AG: Upstream sequences other than AAUAAA are required for efficient messenger RNA 3′-end formation in plants. Plant Cell 2: 1261–1272 (1990).
Aarnes H, Rognes SE: Threonine sensitive aspartate kinase and homoserine dehydrogenase from Pisum sativum. Phytochemistry 13: 2717–2724 (1974).
Bryan JK, Lissik EA, Matthews BF: Changes in enzyme regulation during growth of maize. III. Intracellular localization of homoserine dehydrogenase in chloroplasts. Plant Physiol 59: 673–679 (1977).
Wallsgrove RM, Lea PJ, Miflin BJ: Intracellular localization of aspartate kinase and the enzymes of threonine and methionine biosynthesis in green leaves. Plant Physiol 71: 780–784 (1983).
Azevedo RA, Arana JL, Arruda P: Biochemical genetics of the interaction of the lysine plus threonine resistant mutant Ltr * 1 with opaque-2 maize mutant. Plant Sci 70: 81–90 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ghislain, M., Frankard, V., Vandenbossche, D. et al. Molecular analysis of the aspartate kinase-homoserine dehydrogenase gene from Arabidopsis thaliana . Plant Mol Biol 24, 835–851 (1994). https://doi.org/10.1007/BF00014439
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00014439