Abstract
Co/Ni/Cu/Zn(II) Schiff base complexes with furfural-MAP derived from furfural and 6-methyl-2-aminopyridine have been synthesized and characterized. Quantum chemical parameter calculations of the furfural-MAP and its complexes have been investigated by DFT. The structural characterization have been carried out from their elemental analyses, molar conductance, magnetic, UV–Vis, IR, mass, 1H NMR and EPR. On the basis of spectral data and magnetic measurements, suitable geometries have been proposed for each complex. Redox behaviour of [Cu(II)-(furfural-MAP)2Cl2] has been studied by cyclic voltammetry. Thermal behaviour of [Cu(II)-(furfural-MAP)2Cl2] is consistent with proposed formulation. Powder XRD studies reveal that the compounds are of nanomeric structures. SEM micrograph of furfural-MAP exhibit flake like morphology. NBO, NPA revealed the furfural-MAP to metal charge transfer in complexes. Docking analysis has been performed to identify the interaction between synthesized compounds and active site of EGFR. Antimicrobial activities against various pathogens with reference to known antibiotics and antioxidant activity against standard at variable concentrations exhibit that the metal complexes show remarkable antimicrobial and free radical scavenging activities. In vitro anticancer activity of [Cu(II)-(furfural-MAP)2Cl2] has been studied against human ovarian cancer cells, which exhibit promising anticancer activity. Furthermore, cytotoxicity of [Cu(II)-(furfural-MAP)2Cl2] has been analyzed against L929 cells.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Bioinorganic chemistry focus upon the function of inorganic substances in living systems, including the transport mechanism, speciation and mineralization of inorganic materials and also the use in pharmacotherapy and diagnosis. These substances can be metal ions, composite ions, coordination complexes or inorganic molecules [1, 2]. The chelating property of Schiff bases plays a significant role in its antioxidant activity and this could be helpful in the development of compounds with anticancer activity in transition metal complexes [3]. Density functional theory (DFT) has become an increasingly useful tool for theoretical calculation studies on metal complexes to predict the stereochemical structures [4]. Five membered heterocyclic compounds in arrangement with aromatic moieties perform a noteworthy role in designing a new class of molecules which shows widespread biological behaviour and a dissimilar mechanism of action. Pyridine derivatives hold a diverse array of bioactivities as well as in concert of fundamental roles for physiological functions. These ligands are used in the formation of coordination compounds as medicinal agents [5, 6]. Furfural is a well-known oxy-heterocyclic natural compound used in vegetable oil, petroleum, plastic, rubber, and cosmetic applications [7]. There have been several reports on metal complexes of Schiff bases derived from furfural and its derivatives for their pharmacological and biological applications. In view of the biological importance of parent compounds, here we report synthesis and characterization of furfur-MAP derived from furfural with 6-methyl-2-aminopyridine and its Co(II), Ni(II), Cu(II) and Zn(II)-(furfur-MAP)2Cl2 complexes. DFT, molecular docking, in vitro antioxidant, antimicrobial and cytotoxicity activities have also been carried out in order to ascertain their therapeutic values.
2 Experimental
2.1 Materials
Furfuraldehyde, 6-methyl-2-aminopyridine, Co/Ni/Cu/Zn(II) chloride salts, DMEM, DMSO were obtained from Sigma Aldrich. Sodium bicarbonate, FBS, l-glutamine, methanol and ethanol were bought from Merck. The L929 and PA1 cells were obtained from National Centre for Cell Sciences, Pune, India.
2.2 Characterization techniques
Elemental analysis was obtained using a CHNS Elemental Analyzer—ElementarVario EL III. Systronics MK-509 Digital TDS/conductivity meter was used to measure the conductance. FT-IR spectra were recorded on a Perkin Elmer spectrometer (Model: Spectrum Two) in the range 4000–400 cm−1. Far IR spectra were recorded on a Polytec FIR 30 Far-Infrared Fourier spectrometer using CsI discs. Electronic spectra were recorded on a Perkin Elmer Lambda-35 UV/VIS spectrometer in the range 200–900 nm. Proton NMR spectra were recorded on a BRUKER AVANCE 400 MHz FT-NMR spectrometer employing TMS as an internal standard and DMSO-d6 as solvent. The LC–MS spectra were recorded on a 410 Prostar Binary LC with 500 MS IT PDA Detectors mass spectrometer. The magnetic measurements were studied out using Gouy’s balance and using Pascal’s constant diamagnetic corrections were investigated. The ESR spectrum of CuL2Cl2 complex was recorded in the solid state at RT using Bruker EMX plus ESR Spectrometer and liquid nitrogen temperature using a JES-FA200 ESR Spectrometer. TGA/DSC analysis were performed on a Simultaneous Thermal Analyzer TGDTA under N2 atmosphere. CV measurements were carried out with Princeton Applied Research potentiostat, model VERSASTAT MC. PXRD was recorded on a PANalyticalX’Pert Pro Powder X’Celerator Diffractometer with CuKα (λ = 0.1540 nm) radiation. SEM was carried out on CAREL ZEISS-EVO 18 Scanning Electron Microscope.
2.3 Computational details
Gaussian’09 program [8] has been used to perform the DFT calculations [9] using B3LYP/6-311 ++G (d,p) and B3P86/LANL2DZ basis set [10] for the furfur-MAP and metal(II)-(furfur-MAP)2Cl2 complexes. Natural atomic charges and bonding orbitals are calculated using NBO 3.1 program [11]. Molecular docking was carried out using Auto Dock 4.2 software [12]. The X-ray crystal structure [13] of epidermal growth factor receptor (EGFR) was obtained from RCSB protein data bank [14]. The Lamarckian genetic algorithm [15] was applied to search for protein–ligand interactions and visualization performed using PyMOL [16].
2.4 Synthesis protocols
2.4.1 Schiff base (furfural-MAP)
A solution of furfural (10 mmol, 0.980 g) in MeOH (20 mL) was added dropwise with constant stirring to a methanolic solution of 6-methyl-2-aminopyridine (10 mmol, 1.08 g). The reaction mixture was magnetically stirred and then refluxed for 5 h. The yellow coloured solution obtained was filtered and reduced to half its original volume under boiling water bath. Yellow coloured precipitate was separated out after 1 day. It was recrystallized from EtOH.
(Furfural-MAP): yellowish brown solid; yield: 80%; m.p: 200 °C; Anal. (Cacld.) for C11H10N2O: C, 68.43 (70.96), H, 5.03 (5.37), N, 14.98 (15.05), O, (8.60); LC–MS (m/z): 185.8 [M +]; IR (cm−1): 1664 ν(C=N), 1215ν(C–O), 3080ν(C–H); 1H NMR (DMSO-d6, ppm); –CH=N– (1H, s, 9.01); UV–Vis (λmax, cm−1): 30,959.
2.4.2 Metal complexes [M(II)-(furfural-MAP)2Cl2]
The furfural-MAP (10 mmol) was dissolved in MeOH (20 mL) and metal(II) chlorides (5 mmol) in MeOH (20 mL) was added dropwise with constant stirring and maintained for half an hour. The above complex mixture was magnetically stirred under reflux for 5 h. The solid product formed was filtered off, washed several times with ether and cooled in vacuum desiccator over anhydrous CaCl2.
[Co(II)-(furfural-MAP)2Cl2]: brown solid; yield: 78%; m.p: 220 °C; Anal. (Cacld.) for C22H20N4O2CoCl2: C, 51.39 (52.60), H, 3.56 (3.98), N, 11.25 (11.15), O, (6.37); IR (cm−1): 1650 ν(C=N), 436 ν(M–N), 328 ν(M–Cl); UV–Vis (λmax, nm):548, 627; Ʌc (Ω−1 cm2 mol−1): 10; μeff(B.M): 2.88.
[Ni(II)-(furfural-MAP)2Cl2]: green solid; yield: 81%; m.p: 260 °C; Anal. (Cacld.) for C22H20N4O2NiCl2: C, 52.17 (52.60), H, 3.62 (3.98), N, 11.05 (11.15), O, (6.37); IR (cm−1): 1649 ν(C=N), 461 ν(M–N), 338 ν(M–Cl); UV–Vis (λmax, nm): 458, 545; Ʌc (Ω−1cm2mol−1): 9.
[Cu(II)-(furfural-MAP)2Cl2]: bluish green solid; yield: 76%; m.p: 255 °C; Anal. (Cacld.) C22H20N4O2CuCl2: C, 51.69 (52.12), H, 3.98 (3.94), N, 10.36 (11.05), O, (6.31); LC–MS (m/z): 506.3 [M+]; IR (nm): 1644 ν(C=N), 452 ν(M–N), 335 ν(M–Cl); UV–Vis (λmax, cm−1): 680; Ʌc (Ω−1cm2mol−1): 11; μeff(B.M): 1.86.
[Zn(II)-(furfural-MAP)2Cl2]: colourless solid; yield: 70%; m.p: 260 °C; Anal. (Cacld.) for C22H20N4O2ZnCl2: C, 50.09 (51.93), H, 3.61 (3.93), N, 10.78 (11.01), O, (6.31); IR (cm−1): 1640 ν(C=N), 464 ν(M–N), 338 ν(M–Cl); 1H NMR (DMSO-d6, ppm); –CH=N– (1H, s, 9.11); UV–Vis (λmax, nm): 340; Ʌc (Ω−1 cm2 mol−1): 13.
2.5 Biomedical studies
2.5.1 Antimicrobial activity
The furfural-MAP and its complexes were tested in vitro against the bacterial species, Enterococcus faecalis, Staphylococcus aureus, Escherichia coli and Shigella dysentriae; fungal species, Aspergillus fumigates and Rhizopus stolonifer by Kirby–Bauer disc [17] diffusion method. Vancomycin and clotrimazole were used as standards.
2.5.2 Antioxidant activity
The DPPH is a stable free radical which can accept an electron or hydrogen radical and get changed to a stable, diamagnetic molecule [18]. DPPH (60 μM MeOH) stock solution was prepared. 5, 10, 15 and 20 μg/mL of the test solution (100 μL) was added to a 3.9 mL of DPPH solution. After 20 min incubation at room temperature, the absorbance value was measured against blank solution at 517 nm. The H atom or electron donation capability of the compound was observed from the colour change of purple-colour MeOH solution of DPPH to yellow colour. The experiment was carried out in triplicate and the results were averaged. The % inhibition of free radical production from DPPH was determined using the equation,
2.5.3 Cytotoxic activity
The cytotoxic effect of L929 and PA-1 cells were assessed by MTT assay experiments. The L929 Fibroblast and PA-1 cells were maintained in DMEM. The cell line was cultured in 25 cm2 tissue culture flask with DMEM enhanced with 10% FBS, l-glutamine, sodium bicarbonate and antibiotic solution containing: Penicillin (100 µg/mL), Streptomycin (100 µg/mL), and Amphotericin B (2.5 µg/mL). Cultured cell lines were preserved at 37 °C in a moistened 5% CO2 incubator.
The 2 days old confluent monolayer of cells were suspended in (10%) growth medium, (100 µL) cell suspension (5 × 104 cells/well) was seeded in 96 well tissue culture plate and incubated at 37 °C in a humidified 5% CO2 incubator. 1 mg of complex was weighed accurately and dissolved in 1 mL DMEM using a cyclomixer. The complex solution was filtered through 0.22 µm Millipore syringe filter to ensure the sterility. After 24 h the growth medium was removed, 5% DMEM were 5 times serially diluted by two fold dilution concentrations of 100, 50, 25, 12.5, 6.25 µg in 500 µL of 5% DMEM and each concentration of 100 µL were added in triplicate to the corresponding wells and incubated at 37 °C in a humidified 5% CO2 incubator. Non-treated control cells were also preserved. Entire plate was observed after 24 h of treatment in an inverted phase contrast tissue culture microscope. Any detectable changes in the morphology of cells were considered as indicators of cytotoxicity. 15 mg of MTT was reconstituted in 3 mL PBS until completely dissolved and sterilized by filter sterilization. After 24 h of incubation period, the content in wells were removed and 30 µL of reconstituted MTT solution was added to all test and cell control wells, the plate was gently shaken well, then incubated at 37 °C in a humidified 5% CO2 incubator for 4 h. After the incubation period, the supernatant was removed and 100 µL of MTT solubilization solution of DMSO added and the wells were mixed gently by pipetting up and down in order to solubilize the formazan crystals. The absorbance values were measured by using microplate reader at 540 nm and the cell viability (%) was calculated [19].
3 Results and discussion
3.1 Theoretical studies
3.1.1 Molecular geometry
The optimized molecular geometry of furfural-MAP and its complexes are shown in Fig. 1. The bond length of azomethine C9=N11 is 1.2751 Å in the free furfural-MAP, while the longer bond length of about 1.3089–1.3165 Å were detected in all complexes (Table 1). This can be ascribed to the movement of lone-pair of nitrogen towards vacant orbitals of metal ions, which in turn weakened C9=N11 bond [20]. Moreover the azomethine nitrogen (C=N) bond lengths decrease with an increase in atomic number of the metal ions within the 3d series. The metal-chloride and metal- nitrogen bond lengths in [Cu(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2] are longer than corresponding bond lengths in [Co(II)-(furfural-MAP)2Cl2]and [Ni(II)-(furfural-MAP)2Cl2], the greatest discrepancies being 0.215 Å for M–N between [Zn(II)-(furfural-MAP)2Cl2] and [Ni(II)-(furfural-MAP)2Cl2] and 1.001 Å for M–Cl bond between [Cu(II)-(furfural-MAP)2Cl2] and [Ni(II)-(furfural-MAP)2Cl2]. This shows that the furfural-MAP interacts more strongly with Co(II) and Ni(II) ions than with Cu(II) and Zn(II) ions. In the complexes, the two equivalent hydrogen bonded H10···N17 and H27···N41 contacts resulted in increased stabilization. This is well reflected in the geometrical parameters of pyridine ring in complexes with respect to the furfural-MAP [21]. The observed bond angles (Cl49–M51–Cl50, N11–M51–Cl49, N25–M51–Cl50, N11–M51–N25), suggest tetrahedral geometry for [Co(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2] and square planar geometry [22] for [Ni(II)-(furfural-MAP)2Cl2] and [Cu(II)-(furfural-MAP)2Cl2].
3.1.2 NBO analysis
NBO refers to a set of mathematical algorithms for analyzing electronic wave functions of localized Lewis-like chemical bonds. NBO confirms the of electron transfer from the lone pair of C=N, N11\N25 to metal atoms σ*(M–Cl49\M–Cl50) in complexes having stabilization energy 4.98 kcal/mol. The charge transfer also occurred towards ligand’s furan [LP(1)N11\LP(1)N25 → *(C2–C3)\*(C29–C30)] and pyridine [LP(1)N11\LP(1)N25 → *(C12–C13)\*(C37–C38)] fragments with stabilization energy 0.54 and 1.23 kcal/mol, respectively (Table 2). Comparison of the stabilization energy of these interactions shows that the charge transfer towards the metal atom is more than furan and pyridine rings. The intramolecular C–H···N hydrogen bonding formed by the orbital overlap between the lone pair N17 with *(C9–H10) and N41 with *(C26–H27) antibonding orbital in complexes [23]. Hence the hydrogen bonding interaction leads to an increase in electron density of C–H bonding orbital and thereby weakens the C–H bond. Thus the nature and strength of the intermolecular hydrogen bonding can be analyzed by studying the changes in electron densities in vicinity of C–H···N hydrogen bonds.
3.1.3 Natural population analysis
The NPA has been performed on furfural-MAP and four metal complexes to determine the distribution of electron density. Atomic charge distribution of furfural-MAP and its complexes show that all carbon atoms are negative except C1, C2, C9, C12, C16, C26, C28, and C31; those carbon atoms bonded to N or O atoms. The magnitudes of atomic charges on all hydrogen atoms are complexes are positive, charge ranging from 0.1921 e to 0.2517 e. The natural charges on Cl49 (− 0.5110 e) and Cl50 (− 0.4088 e) attached to the metal ions are extremely lower than that of the formal − 1 charge on a free chloride ion due to their coordination with the central metal ions, which increase in the order Ni–Cl < Co–Cl < Cu–Cl < Zn–Cl. The transition metal ions in complexes are formally in the second oxidation state, here the computed natural charges Co+0.4289, Ni+0.3025, Cu+0.6323and Zn+1.1089 are considerably lower than +2 manifest metal-to-ligand back donations in these complexes. Moreover, the magnitude of negative charge on the azomethine nitrogen (N11 and N25, − 0.4297e) donor atom of furfural-MAP appreciably increased in complexes. This implies that metal to ligand back donation of electrons occurs in complexes via azomethine nitrogen atom [24].
3.1.4 Frontier molecular orbital analysis
Spatial distribution of molecular orbitals, especially those of HOMO and LUMO, are good indicators of electron transport in molecular systems. Molecules having small HOMO–LUMO energy gap are generally associated with high chemical reactivity and low kinetic stability [25]. In the furfural-MAP, HOMO density is spread throughout the molecule. A highly delocalized HOMO indicates that the electrons can more readily move around the molecule and hence an enhanced intramolecular charge transfer. On the other hand, the LUMO localizes on pyridine and furan rings indicating the presence of favorable atomic centers within furfural-MAP for possible nucleophilic attacks. The HOMO and LUMO orbitals of furfural-MAP and its complexes are shown in Fig. 2. In the complexes HOMO density is located the metal, azomethine nitrogen and chlorine moieties whereas LUMO maps have manifested the charge distribution on the entire molecule. This result reveals the ultimate charge transfer from the electron-donor groups to electron-acceptor groups through coordination. Furthermore, the computations show that frontier molecular orbital energy gap follows the order of [Cu(II)-(furfural-MAP)2Cl2] = [Ni(II)-(furfural-MAP)2Cl2] (− 1.1017 eV) < [Zn(II)-(furfural-MAP)2Cl2] (− 3.9158 eV) < [Co(II)-(furfural-MAP)2Cl2] (− 4.1828 eV) < furfural-MAP (− 4.3174 eV). The lowering of the HOMO–LUMO band gap in [Cu(II)-(furfural-MAP)2Cl2] and [Ni(II)-(furfural-MAP)2Cl2] is essentially a consequence of the large stabilization of the LUMO due to the strong electron-accepting ability of the electron-acceptor group. It is worth noting that the charge transfer interaction through C–H···N hydrogen bonding results in substantial ground state donor–acceptor mixing. Consequently, an electron density transfer occurs from the aromatic part in the electron-donor side to its electron-withdrawing part.
3.1.5 Molecular docking
Molecular docking with the crystal structure of EGFR (PDB code: 1M17) has been undertaken to identify the level of antitumour activities of furfural-MAP and complexes over ovarian cancer in which epidermal growth factor receptor [26] is highly expressed. Pictorial representations of best possible binding sites of EGFR with synthesized compounds are shown in Fig. 3. The results of the docking studies (Table S1) show that the compounds interacted favourably with the active binding sites of the proteins. Calculated free energy of binding for [Cu(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2] in the receptor binding site was − 5.56 kcal/mol and − 5.57 kcal/mol respectively. It is interesting that there is one kind of hydrogen bond that is formed by the furan oxygen atom of [Zn(II)-(furfural-MAP)2Cl2] and the amino hydrogen of TYR 1106. In contrast, there are two kinds of hydrogen bonds in the binding model of [Cu(II)-(furfural-MAP)2Cl2]. One kind was formed by the furan oxygen atom and hydroxyl group of ALA 866 and the other was composed of amino hydrogen of CYS 919 and azomethine nitrogen. In comparison to [Zn(II)-(furfural-MAP)2Cl2] protein interactions, [Cu(II)-(furfural-MAP)2Cl2] may also form hydrophobic interactions with GLU 917, LEU 840 and VAL 848. Visual analysis of the results showed that [Ni(II)-(furfural-MAP)2Cl2] as whole was focused in the groove of the protein 1M17 forming 2-H bonds with VAL 848 and ILE 1084. The docking interactions of [Co(II)-(furfural-MAP)2Cl2] with 1M17 showed the binding energy of − 5.46 kJ/mol. This interaction is favored by the formation of H-bond with LYS 826 and THR 916 and and hydrophobic interactions with ASP 857 and LEU 902.
3.2 Experimental studies
Newly synthesized furfural-MAP and its [M(II)-(furfural-MAP)2Cl2] showed 1:2 metal‐to‐ligand stoichiometry for all complexes. They are readily soluble in EtOH and DMF. The purity was examined by TLC. The low molar conductance values (Sect. 2.3) of [M(II)-(furfural-MAP)2Cl2] in DMSO solution at 10−3 M exhibit the non-electrolytic behaviour [27].
3.2.1 Infrared spectra
The IR spectral data of furfural-MAP and its complexes are given in Sects. 2.2 and 2.3. The IR spectrum of furfural-MAP show a strong band at 1664 cm−1 is assigned to stretching vibration of ν(C=N) vibrations [28]. During complex formation this band was immediately shifted to the lower region 1639–1650 cm−1 indicate coordination of azomethine nitrogen atom to metal ion. The medium intensity bands appeared in the metal complexes at 436, 461, 452 and 464 cm−1 are attributed to ν(Co–N), ν(Ni–N), ν(Cu–N) and ν(Zn–N) bands, respectively [29]. In addition to this, presence of medium intensity bands in the furfural-MAP at 1215 and 3080 cm−1 are assigned to ν(C–O) and ν(C–H) stretching vibration modes of furan [30]. In the complexes, the bands agreeing to ν(C–O) and ν(C–C) of furan ring seemed at the same position as observed in the furfural-MAP and also lack of ν(M–O) band reinforced the non-participation of furan oxygen atom in coordination. Far IR spectra of the [M(II)-(furfural-MAP)2Cl2] showed new bands at 328, 333, 335 and 338 cm−1 may be ascribed to ν(Co–Cl), ν(Ni–Cl), ν(Cu–Cl) and ν(Zn–Cl) vibrations, respectively.
3.2.2 1H NMR spectra
The 1H NMR spectrum of furfural-MAP, revealed a significant azomethine proton [31] signal (s, CH=N) at 9.01 δ, ppm where as multiplets in the range 6.42–6.58δ, ppm (m, ArH) attributed to aromatic ring protons [32]. In [Zn(II)-(furfural-MAP)2Cl2] azomethine proton signal was observed at 9.10 δ ppm. This deshielding of CH=N proton is due to coordination of azomethine nitrogen with the metal atom.
3.2.3 Mass spectra
The mass spectrum of furfural-MAP shows a molecular ion peak at m/z 186.48 consistent with the suggested molecular formula of the Schiff base. The mass fragmentation peaks seemed at m/z 117.2 and 92.1 are due to loss of [C4H4O] and [C6H6N] moieties from the molecular ion peaks. The molecular ion peaks of the [M(II)-(furfural-MAP)2Cl2] have been taken into account to support their suggested molecular formulae. The molecular ion peak of [Cu(II)-(furfural-MAP)2Cl2] observed at m/z 506.3 agrees well with 1:2 (M:L) ratio stoichiometry. Elemental data coincides well with experimental mass spectral data values for the newly synthesized compounds.
3.2.4 Electronic spectra and magnetic data
The electronic spectrum of furfural-MAP displayed an absorption band at 323 nm may be assigned to n–π* transition which is the characteristic of C=N group. The position of this spectral band was shifted to higher wavelength region in the spectra of [M(II)-(furfural-MAP)2Cl2] suggesting the coordination of furfural-MAP to the metal ions.
The [Co(II)-(furfural-MAP)2Cl2] displayed two bands at 548 and 627 nm assignable to 4A2g → 4T1g(F) and 4A2g → 4T1g(P) transitions in tetrahedral geometrical environment. The magnetic moment value of 2.88 BM further corroborates this observation [33]. The observed absorption bands of [Ni(II)-(furfural-MAP)2Cl2] at 458 and 545 nm corresponds to 4A1g → 4A2g and 4A1g → 4B1g transitions respectively, characteristic of a square planar geometrical environment [34]. The broad absorption band detected in the electronic spectrum of [Cu(II)-(furfural-MAP)2Cl2] at 680 nm may be accredited to 2Eg → 2T2g transition designate square planar configuration. The observed magnetic moment value of [Cu(II)-(furfural-MAP)2Cl2] is 1.86 B.M., which confirms its geometry. [Zn(II)-(furfural-MAP)2Cl2] is diamagnetic and has completely filled d10 configuration, therefore it would have tetrahedral geometry [35].
3.2.5 EPR spectrum
The ESR spectrum of [Cu(II)-(furfural-MAP)2Cl2] is shown in Fig. 4. The analysis of spectrum gives g║ = 2.193 and g⊥ = 2.035. The relation g║ > g⊥ > ge, observed for the complex indicated that one the unpaired electron is most likely in the dx2 − y2 orbital [36]. The g║ value of [Cu(II)-(furfural-MAP)2Cl2] is less than 2.3, shows noticeable covalent character for the metal–ligand bond [37]. The geometric parameter, G is a measure of degree of exchange interactions and is calculated from expression G = (g║ − 2)/(g⊥ − 2). If G > 4, the spin-exchange interaction is negligible and if it is less than 4 considerable spin-exchange interaction prevails. The calculated G value of [Cu(II)-(furfural-MAP)2Cl2] is 5.51, indicates the exchange coupling effects are not operative in the complex. The [Cu(II)-(furfural-MAP)2Cl2] at liquid nitrogen temperature shows four peaks and are obviously due to the coupling of the electron spin of the 63Cu nucleus (I = 3/2). The peaks are broad and have the appearance of well-resolved triplets. The breadth and triplet appearance can be attributed to hyperfine splitting by the nitrogen atom (I = 1) of the ligand. Based on this observation, a square planar geometry is proposed for the complex. The EPR study of the [Cu(II)-(furfural-MAP)2Cl2] has provided supportive evidence to the conclusion obtained on the basis of electronic spectrum and magnetic moment value.
3.2.6 Cyclic voltammetry
The electrochemical response of [Cu(II)-(furfural-MAP)2Cl2] was analyzed in DMSO solution at a scan rate of 100 mV s−1 in the potential range + 1.5 to − 1.5 V. Cyclic voltamogram of [Cu(II)-(furfural-MAP)2Cl2] (supplementary Fig. S1) exhibits the reduction peak potential at Epc = − 0.112 V with the corresponding oxidation peak potential Epa = − 0.684 V, which indicates the formation of Cu(II)/Cu(I) couple. The peak-to-peak separation (ΔEp = 0.572 V) indicates quasi-reversible one electron transfer process [38, 39] for [Cu(II)-(furfural-MAP)2Cl2].
3.2.7 Thermal analysis
The thermal stabilities of furfural-MAP and [Cu(II)-(furfural-MAP)2Cl2] (Fig. 5) were examined in N2 atmosphere at a heating rate of 10 °C/min, and the mass loss was measured from 30 to 900 °C. The TGA curve of furfural-MAP is stable up to 190 °C and no weight loss occurs before this temperature region. The decomposition occurred in a single step, starts from 190 °C and ends at 400 °C. An almost horizontal thermal curve [40] was observed above 400 °C. The thermogram of [Cu(II)-(furfural-MAP)2Cl2] shows decomposition in two steps. There is no weight loss up to the temperature of ~ 200 °C, which infers the absence of lattice and coordinated water molecule. The decomposition, starts from 220 °C and ends at 245 °C with an endothermic DSC peak in the range 230–245 °C. It is attributed to the loss of chloride ions this step bringing the weight loss of 15.48%. The resultant complex underwent second stage degradation in the range of 350–450 °C with a weight loss of 69% as a result of decomposition of furfural-MAP molecule. The [Cu(II)-(furfural-MAP)2Cl2] is being in the form of its CuO as above 450 °C. The ligand and its metal complex structures are shown in Fig. 6.
3.3 Nanocrystalline studies
3.3.1 Powder XRD
Powder XRD pattern of furfural-MAP and its complexes were recorded over the 2θ = 10–80◦ and its patterns are shown in Fig. 7. The furfural-MAP, [Cu(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2] displays a sharp crystalline peaks indicating their crystalline nature as Nano regime, while [Co(II)-(furfural-MAP)2Cl2] and [Ni(II)-(furfural-MAP)2Cl2] do not exhibit well defined crystalline peak indicating their amorphous behaviour. The furfural-MAP, [Cu(II)-(furfural-MAP)2Cl2], and [Zn(II)-(furfural-MAP)2Cl2] have a calculated average crystallite size of 43, 52, and 46 nm respectively, indicating that the obtained complexes are in nanocrystalline state.
3.3.2 SEM
The SEM analysis has been carried out to evaluate the surface morphology of furfural-MAP and metal complexes (supplementary Fig. S2). The micrographical images of furfural-MAP and metal complexes differ significantly due to coordination of metal ions with the donor sites in the furfural-MAP. Also SEM micrographs of the metal complexes revealed that the surface morphology of metal complexes changes by changing the metal ions. The SEM images of furfural-MAP and [Cu(II)-(furfural-MAP)2Cl2] indicate non-uniform flake-like structures [41]. The micrographs of [Co(II)-(furfural-MAP)2Cl2] depicts the presence of cauli-flower like morphology, on the other hand [Ni(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2] are faceted with spine like morphology [42].
3.4 Biomedical studies
3.4.1 Antimicrobial activity
The in vitro antimicrobial activity was assessed by measuring the zone of inhibition method for the synthesized compounds. The results (supplementary Fig. S3, Table S2) show that the activity follows the order [Ni(II)-(furfural-MAP)2Cl2] < [Cu(II)-(furfural-MAP)2Cl2] < [Zn(II)-(furfural-MAP)2Cl2] < [Co(II)-(furfural-MAP)2Cl2] < furfural-MAP. The metal complexes are found to be more toxic against the tested bacterial species than the furfural-MAP. Complexation reduces polarity of metal ion by the partial sharing of its positive charge with donor groups in complexes. This in turn increases the lipophilic nature of the central metal atom which eventually favors its permeation through the lipid layer of the cell membrane. The [Ni(II)-(furfural-MAP)2Cl2] and [Cu(II)-(furfural-MAP)2Cl2] were seemed to be more toxic towards both gram positive and gram negative bacteria than the other metal complexes; this may be due to presence of Ni(II) and Cu(II) ions. The results of fungicidal screening activity show that, the furfur-MAP has comparable activity with its complexes against the two types of tested fungi. The mode of action may involve the formation of H-bond through the azomethine nitrogen atom with the active centers of the cell constituents, resulting in interference with the normal cell process [43]. These results could pave the way for the development of next generation antibiotics for therapeutic purpose.
3.4.2 Antioxidant activity
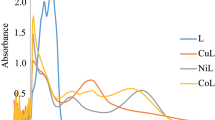
The radical scavenging effect of furfural-MAP and complexes are shown as concentration dependent curve in Fig. S4, Supplementary. The antioxidant potency has been assessed by DPPH method using L-ascorbic acid as standard. Metal complexes showed higher activity than the ligand which is mainly due to the chelation of organic ligand with the metal ion. [Ni(II)-(furfural-MAP)2Cl2] and [Cu(II)-(furfural-MAP)2Cl2] exhibited best scavenging activity, while [Zn(II)-(furfural-MAP)2Cl2] has shown moderate activity. The variation in the antioxidant activity of metal complexes is due to coordination environment and redox properties [44, 45].
3.4.3 Cytotoxic activity
The cytotoxicity [Cu(II)-(furfural-MAP)2Cl2] on PA1(Ovarian cancer) and L929(fibroblast) cell lines has been evaluated using MTT assay [46, 47]. The results indicate that [Cu(II)-(furfural-MAP)2Cl2] have the cytotoxic effects at various concentrations (6.5 to 100 μg/mL). The cell viability of [Cu(II)-(furfural-MAP)2Cl2] against PA1 and L929 cells (supplementary Fig. S5) illustrated that increase in concentration of [Cu(II)-(furfural-MAP)2Cl2] adversely decreased the viability of the treated cell lines. Also, the highest concentration applied showed the highest decrease in cell viability in both cell lines. The cell viability of L929 cells were greater than 51% for all concentrations tested, indicates that the [Cu(II)-(furfural-MAP)2Cl2] can be used as a safe compound for therapeutic biomedical applications. Both PA1 and L929 cells showed no statistically significant difference in cell viability when treated with a concentration ranging from 6.25 to 12.5 μg/mL. On increasing the concentration above12.5 μg/mL, resulted a drastic change in the morphological characteristics of tested cell lines. Cells began to shrink and lose their capacity to adhere the surface of cultivation plate. Cells become rounded and completely floated in comparison to the control morphology at the highest applied concentration. Furthermore the [Cu(II)-(furfural-MAP)2Cl2] shows > 51% viable L929 cells for all concentrations tested indicates that the [Cu(II)-(furfural-MAP)2Cl2] can be used as a safe compound for therapeutic applications. These results suggest that [Cu(II)-(furfural-MAP)2Cl2] is able to develop an inhibitory effect on the proliferation of the PA1 cell line. Furthermore, the 50% inhibitory concentration at 48 h exposure for cisplatin was 42 μg/mL compared with 91.42 μg/mL for [Cu(II)-(furfural-MAP)2Cl2].
4 Conclusions
The stoichiometry and bonding modes of furfural-MAP and metal complexes have been deduced from the relevant features of both theoretical and experimental results. The geometrical structures are found to be square planar for [Ni(II)-(furfural-MAP)2Cl2] and [Cu(II)-(furfural-MAP)2Cl2] and tetrahedral for [Co(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2]. Thermal stability and decomposition stages were observed by thermal studies. The crystalline nature, crystallite size and morphology have been obtained from PXRD and SEM analysis. NBO analysis clearly demonstrates the formation of H-bonded interactions. Atomic charge calculations indicate that charge transfer has been taking place from the furfural-MAP to the metal as well as back-donation from the metal ion to the donating sites. Molecular docking studies indicate greater interactions of EGFR with [Cu(II)-(furfural-MAP)2Cl2] and [Zn(II)-(furfural-MAP)2Cl2] evidenced by best binding energy. The [Cu(II)-(furfural-MAP)2Cl2] compound showed a good cytotoxic effect when compared with the other prepared compounds.
References
Liu X, Hamon JR (2019) Recent developments in penta, hexa and heptadentate Schiff base ligands and their metal complexes. Coord Chem Rev 389:94–118
Nakano K, Kobayashi K, Nozaki K (2011) Tetravalent metal complexes as a new family of catalysts for copolymerization of epoxides with CO2. J Am Chem Soc 133(28):10720–10723
Ghosh D, Choudhury ST, Ghosh S, Mandal AK, Sarkar S, Ghosh A, Saha KD, Das N (2012) Nanocapsulated curcumin: oral chemopreventive formulation against diethylnitrosamine induced hepatocellular carcinoma in rat. Chem Biol Interact 195(3):206–214
Jafari M, Salehi M, Kubicki M, Arab A, Khaleghian A (2017) DFT studies and antioxidant activity of Schiff base metal complexes of 2-aminopyridine. Crystal structures of Co(II) and Zn(II) complexes. Inorg Chim Acta 462:329–335
Kaplancikli ZA, Turan-Zitouni GA, Ozdemir Revial G (2008) New triazole and triazolothiadiazine derivatives as possible antimicrobial agents. Eur J Med Chem 43(1):155–159
Rezaeivala M, Keypour H (2014) Schiff base and non-Schiff base macrocyclic ligands and complexes incorporating the pyridine moiety; the first 50 years. Coord Chem Rev 280:203–253
Wu SN, Chang HD (2006) Diethyl pyrocarbonate, a histidine-modifying agent, directly stimulates activity of ATP-sensitive potassium channels in pituitary GH-3 cells. Biochem Pharmacol 71(5):615–623
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa JH, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, Revision A.02. Gaussian Inc., Wallingford
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98(7):5642–5648
Hay PJ, Wadt WR (1985) Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J Chem Phys 82(1):99–310
Foster JP, Weinhold F (1980) Natural hybrid orbitals. J Am Chem Soc 102(24):7211–7218
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Auto Dock 4 and Auto Dock Tools 4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Fricker J (2006) Tyrosine kinase inhibitors: the next generation. Lancet Oncol 7(8):621
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semi empirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152
The PyMOL Molecular Graphics System (2009) Version 1. 5.0.4, Schrodinger, LLC
Bauer AW, Kirby WMM, Sherris JC, Truck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Lone IH, Khan KZ, Fozdar BI, Hussain F (2013) Synthesis antimicrobial and antioxidant studies of new oximes of steroidal chalcones. Steroids 78(9):945–950
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65(1–2):55–63
Smith Brian (1998) Infrared spectral interpretation, 1st edn. CRC Press, New York
Klein RA (2006) Modified van der Waals atomic radii for hydrogen bonding based on electron density topology. Chem Phys Lett 425(1):128–133
Ghogomu JN, Nkungli NK (2016) A DFT study of some structural and spectral properties of 4-methoxyacetophenone thiosemicarbazone and its complexes with some transition metal chlorides: potent antimicrobial agents. Adv Chem 2016:1–15
Kulkarni AD, Truhlar DG (2011) Performance of DFT and Møller–Plesset second-order perturbation theory for structural parameters in complexes of Ru. J Chem Theory Comput 7(7):2325–2332
Karnan M, Balachandran V, Murugan M, Murali MK, Nataraj A (2013) Vibrational FT-IR and FT-Raman spectra, NBO, HOMO–LUMO, molecular electrostatic potential surface and computational analysis of 4-(trifluoromethyl)benzylbromide. Spectrochim Acta A 116:84–95
Fleming I (2010) Molecular orbitals and organic chemical reactions, 1st edn. Wiley, Chichester
Mahmoud WH, Omar MM, Sayed FN, Mohamed GG (2018) Synthesis, characterization, spectroscopic and theoretical studies of transition metal complexes of new nano Schiff base derived from l-histidine and 2-acetylferrocene and evaluation of biological and anticancer activities. Appl Organomet Chem 32(7):4386–4390
Geary J (1971) The use of conductivity measurements in organic solvents for the characterization of coordination compounds. Coord Chem Rev 7(1):81–122
Nawar N, Hosny NM (2000) Synthesis, spectral and antimicrobial activity studies of o-aminoacetophenone, o-hydroxybenzoylhydrazone complexes. Transition Met Chem 25(1):1–8
El-Baradie KY, El-Wakiel NA, El-Ghamry HA (2014) Synthesis, characterization and corrosion inhibition in acid medium of l-histidine Schiff base complexes. Appl Organomet Chem 29(3):117–125
Shakir M, Abbasi A, Khan AU, Khan SN (2011) Synthesis and spectroscopic studies on the Schiff base ligand derived from condensation of 2-furaldehyde and diaminobenzidene, and its complexes with Co(II), Ni(II), Cu(II) and Zn(II): comparative DNA binding studies of ligand and its Cu(II) and Zn(II) complexes. Spectrochim Acta A 78(1):29–35
Refat MS, El-Sayed MY, Adam AMA (2013) Cu(II), Co(II) and Ni(II) complexes of new Schiff base ligand: synthesis, thermal and spectroscopic characterizations. J Mol Struct 1038:62–72
Ali OAM (2014) Corrigendum to “Experimental and theoretical methods of (E)-3- methoxy-2-[(p-tolylimino)methyl]phenol. Spectrochim Acta A 132:52–60
Dutta RI, Syamal A (1993) Elements of mageneto chemistry, 2nd edn. EWP, New Delhi
Sallam SA (2006) Binuclear Cu(II), Ni(II) and Co(II) complexes with N2O2 chromophores of glycylglycine Schiff bases of acetylacetone, benzoylacetone and thenoyltrifluoroacetone. Transition Met Chem 31(1):46
Todor D, Carmay LJ (2000) Tetrahedral vs octahedral zinc complexes with ligands of biological interest: a DFT/CDM study. Am Chem Soc 122(45):11146–11153
Muller J, Schubi D, Mossmer CM, Strahle J, Weser U (1999) Structure-function correlation of Cu(II) and Cu(I) di-Schiff-base complexes during the catalysis of superoxide dismutation. Inorg Chim Acta 75(1):63–69
Kivelson D (1997) Publications of Daniel Kivelson. J Phy Chem B 101(43):8631–8634
Hathaway BJ, Billing DE (1970) The electronic properties and stereochemistry of mono-nuclear complexes of the Cu(II) ion. Coord Chem Rev 5(2):143–207
Bard AJ, Faulkner LR (1980) Electrochemical methods. Wiley, New York
Emam SM (2017) Spectral characterization, thermal and biological activity studies of Schiff base complexes derived from 4,4′-methylenedianiline, ethanol amine and benzyl. J Mol Struct 1134:444
Arish D, Nair MS (2011) Synthesis, spectroscopic, antimicrobial, DNA binding and cleavage studies of some metal complexes involving symmetrical bidentate N, N donor Schiff base ligand. Spectrochim Acta A 82(1):191–199
Joseyphus RS, Nair MS (2008) Synthesis, characterization and antimicrobial activity of transition metal complexes with the Schiff base derived from imidazole-2-carboxaldehyde and glycylglycine. J Coord Chem 62(2):319–327
Raja D, Bhuvanesh NSP, Natarajan K (2012) A novel water soluble ligand bridged Co(II) coordination polymer of 2-oxo-1,2-dihydroquinoline-3-carbaldehyde (isonicotinic) hydrazone: evaluation of the (DNA) binding, protein interaction, radical scavenging and anticancer activity. Dalton Trans 41(15):4365
Manimohan M, Pugalmani S, Sithique MA (2019) Biologically active novel N, N, O donor tridentate water soluble hydrazide based O-carboxymethyl chitosan Schiff base Cu (II) metal complexes: synthesis and characterisation. Int J Biol Macromol 136:738–754
Tverdova NV, Pelevina ED, Giricheva NI, Girichev GV, Kuzmina NP, Kotova OV (2011) Molecular structure of N,N′-o-phenylene-bis(salicylideneaminato)copper(II) studied by gas-phase electron diffraction and quantum-chemical calculations. Struct Chem 22:441
Inan A, Ikiz M, Tayhan SE, Bilgin S, Genç N, Sayın K, Ceyhan G, Kose M, Dag A, Ispir E (2018) Antiproliferative, antioxidant, computational and electrochemical studies of new azo-containing Schiff base ruthenium (II) complexes. New J Chem 42(4):2952–2963
Pravin N, Kumaravel G, Senthilkumar R, Raman N (2017) Mixed ligand Cu(II) complexes containing a Schiff base and diimine. Appl Organomet Chem 31(10):3714–3739
Zhang P, Huang H (2018) Future potential of osmium complexes as anticancer drug candidates, photosensitizers and organelle-targeted probes. Dalton Trans 47(42):14841–14854
Acknowledgements
Mrs. Liji John acknowledges the University of Kerala, Thiruvananthapuram, Kerala, India for providing the University-JRF. We are thankful to SAIF, IIT-Bombay; CECRI, Karaikudi; CLIF, University of Kerala; M. S. University, Tirunelveli; St Joseph`s College (Autonomous), Tiruchirappalli; N. I. University, Kumaracoil; Athmic biotech solutions, Kalliyoor; Biogenix research center for molecular biology & applied sciences, Poojappura, India for extending the research activities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
John, L., Joseyphus, R.S. & Joe, I.H. Biomedical application studies of Schiff base metal complexes containing pyridine moiety: molecular docking and a DFT approach. SN Appl. Sci. 2, 500 (2020). https://doi.org/10.1007/s42452-020-2274-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2274-6