Abstract
The high-throughput - next generation sequencing (HT-NGS) technologies are currently the hottest topic in the field of human and animals genomics researches, which can produce over 100 times more data compared to the most sophisticated capillary sequencers based on the Sanger method. With the ongoing developments of high throughput sequencing machines and advancement of modern bioinformatics tools at unprecedented pace, the target goal of sequencing individual genomes of living organism at a cost of $1,000 each is seemed to be realistically feasible in the near future. In the relatively short time frame since 2005, the HT-NGS technologies are revolutionizing the human and animal genome researches by analysis of chromatin immunoprecipitation coupled to DNA microarray (ChIP-chip) or sequencing (ChIP-seq), RNA sequencing (RNA-seq), whole genome genotyping, genome wide structural variation, de novo assembling and re-assembling of genome, mutation detection and carrier screening, detection of inherited disorders and complex human diseases, DNA library preparation, paired ends and genomic captures, sequencing of mitochondrial genome and personal genomics. In this review, we addressed the important features of HT-NGS like, first generation DNA sequencers, birth of HT-NGS, second generation HT-NGS platforms, third generation HT-NGS platforms: including single molecule Heliscope™, SMRT™ and RNAP sequencers, Nanopore, Archon Genomics X PRIZE foundation, comparison of second and third HT-NGS platforms, applications, advances and future perspectives of sequencing technologies on human and animal genome research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The completion of the first human genome drafts (Yamey 2000) was just a start of the modern DNA sequencing era which resulted in further invention, improved development toward new advanced strategies of high-throughput DNA sequencing, so called the “high-throughput next generation sequencing” (HT-NGS). These developed HT-NGS strategies addressed our anticipated future needs of throughput sequncing and cost, in a way which enabled its potential multitude of current and future applications in mammalian genomic research. Additionally in these advanced laboratory methodologies, a scope of new generation of bioinformatics tools has further emerged as an essential prerequisite to accommodate further strategic development and improvement of output results. The HT-NGS is one of the great challenges of today’s genomic research. For the future direction, we need the in-depth genome sequence information and analysis for most of the mammals, including human to fully understand genome variation of economic traits, genetic susceptibility to diseases, and pharmacogenomics of drug response. The leading genome research centers and scientists have publicly recognized that these are the core enabling goals for the next decade genomics research. The National Human Genome Research Institute (NHGRI) has echoed this need through its vision for genomics research (Collins et al. 2003). The NHGRI has categorized new sequencing approaches into those that offer near-term and revolutionary benefits with a 100-fold cost reduction per base pair (bp) within the next five years. To extend the near-term, i.e., of within the next 5–10 years, the revolutionary benefits should advance the field with a 10,000-fold cost reduction per base pair which in turn to attain the “US$ 1000 genome”.
Year 2011 is celebrated as the 10th anniversary since the human genome was first sequenced (www.nature.com/natureconferences/hg10years/index.html). During this period, tremendous success has been achieved in the fields of decoding of human genome, technological advancement of new era of human genome applications, toward personalized genomes and discovery of rare variants, leveraging genome sequencing to impact on cancer researches and mammalian evolution and population structure. The past decade has witnessed a revolution in the field of human genomics research. Today, a more global approach is being embraced which has not only given a rise to the field of systems biology, but has also touched all areas of biological and medical research, as well as bringing them closer together and blurring the lines that previously defined them as individual disciplines of research. The horizons and expectations have broadened due to the technological advances in the field of genomics, especially the HT-NGS and its wide range of applications such as: chromatin immunoprecipitation coupled to DNA microarray (ChIP-chip) or sequencing (ChIP-seq), RNA sequencing (RNA-seq), whole genome genotyping, de novo assembling and re-assembling of genome, genome wide structural variation, mutation detection and carrier screening, detection of inherited disorders and complex human diseases, DNA library preparation, paired ends and genomic captures, sequencing of mitochondrial genome and personal genomics (for the detailed description, see: Table 2). Besides the advancement of sequencing techniques, the past decade will be remembered as the decade of the genome research. Since the publications of first composite genomes of human (Lander et al. 2001; Venter et al. 2001) - many draft genomes from other organisms have been published (www.ensembl.org/info/about/species.html). The speed with which new genomes can now be sequenced has been facilitated by the development of potential HT-NGS technologies and assembly methods. It is now possible to assemble de novo a large genome, a good example can be derived from the recent genome assembly of the giant panda (Li et al. 2010b) which utilized the only short reads provided by next-generation DNA sequencing.
First generation DNA sequencers
Historically in 1975, the Sanger introduced the concept of DNA sequencing method in his pioneered Croonian lecture (Sanger 1975) and later on, published a rapid method for determining sequences in DNA by primed synthesis with DNA polymerase (Sanger and Coulson 1975). In the year of 1977, two landmark articles for DNA sequencing were published, i.e., the Frederick Sanger’s enzymatic dideoxy DNA sequencing technique based on the chain-terminating dideoxynucleotide analogues (Sanger et al. 1977) and the Allan Maxam and Walter Gilbert’s chemical degradation DNA sequencing technique in which terminally labeled DNA fragments were chemically cleaved at specific bases and separated by gel electrophoresis (Maxam and Gilbert 1977). These two prominent elite laboratories were responsible for the introduction of the first automated DNA sequencers led by Caltech (Smith et al. 1986), which was subsequently commercialized by Applied Biosystems (ABI), the European Molecular Biology Laboratory (EMBL) (Ansorge et al. 1986, 1987) and Pharmacia-Amersham, later General Electric (GE) healthcare. This refinement and commercialization of the sequencing method led to its broad dissemination throughout the global research community.
In the first automated fluorescent DNA sequencing equipment, a complete gene locus for the hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene was sequenced, using for the first time the paired-end sequencing approach (Edwards et al. 1990). In 1996, ABI introduced the first commercial DNA sequencer that utilized a slab gel electrophoresis by the ABI Prism 310. Two years later, the considerable labor of pouring slab gels was replaced with automated reloading of the capillaries with polymer matrix by ABI Prism 3700 with 96 capillaries. This automated DNA sequencer was successfully utilized in the sequencing of the first human genome in 2003 taking into account 13-years of efforts of the human genome project consortium, and with an estimated cost of $2.7 billion. In the following years, another landmark was achived by the DNA sequencing of the first small phage genome (5386 bases in length) and sequencing of the human genome of upto ∼ 3 billion bases (Lander et al. 2001; Venter et al. 2001). It is remarkable that such progress has been made using methods that are refinements of the basic ‘dideoxy’ method introduced by Sanger in 1977.
Birth of HT-NGS
In 2000, Jonathan Rothberg founded 454 Life Sciences, which further developed the first commercially available NGS platform, the GS 20. The GS instrument was introduced in 2005, developed by 454 Life Sciences (www.454.com), as the first NGS system on the market. The developed technique was successfully validated by combining single-molecule emulsion PCR with pyrosequencing (shotgun sequencing procedure) of the entire 580 069 bp of the Mycoplasma genitalia genome at 96% coverage and 99.96% accuracy in a single GS 20 run (Margulies et al. 2005). In the following years, Roche applied science acquired 454 Life sciences and extended further the new version of the 454 instrument, i.e., the GS FLX titanium. Sharing the same technological principle in both GS 20 and GS FLX titanium, the flow cell is referred to as a "picotiter well" plate, which is made from a fused fiber-optic bundle. On a separate front, single-molecule PCR in microcompartments consisting of water-in-oil emulsions was also developed by Roche HT-NGS platform (Tawfik and Griffiths 1998). In general, the principle of pyrosequencing technique is based on the “sequencing by synthesis”. It differs from Sanger sequencing because, it depends on the detection of pyrophosphate release on nucleotide incorporation, rather than chain termination with dideoxynucleotides. The technique was developed by joint efforts of Swedish group (the teams of M. Ronaghi, M. Uhlen, and P. Nyren) in Stockholm (Ronaghi et al. 1996). They first described a sequencing approach based on chemiluminescent detection of pyrophosphate released during polymerase-mediated deoxynucleoside triphosphate (dNTP) incorporation (Nyren et al. 1993, Nyren 2007) and the real-time DNA sequencing, utilizing this release of pyrophosphate detection (Ronaghi et al. 1998). In pyrosequencing the DNA synthesis is performed within a complex reaction that includes ATP sulfurylase and luciferase enzymes and adenosine 5′ phosphosulfate and luciferin substrates in such a way that, the pyrophosphate group releases upon addition of a nucleotide, resulting in the production of detectable light.
The HT-NGS techniques, which are new opportunities and a great impact on mammalian genomics research were selected as the methods of the year in 2007 (Schuster et al. 2008). However, the road to gain the acceptance of these novel technologies was not an easy one. The first step of the HT-NGS technique consisted in detecting the next added fluorescently labeled base (reversible terminator) in the growing DNA chain by means of a sensitive CCD camera. This was performed on a large number of DNA samples in parallel, attached either to a planar support or to beads, on DNA chips, minimizing reaction volumes in a miniaturized microsystem. In the next step the terminator was converted into a standard nucleotide and the dye was removed. This cycle and the process were repeated to determine the next base in the sequence. The principle described in this application is in part very quasi to that used today in the so-called next-generation devices, commercialized by Roche, Illumina-Solexa, ABI, Helicos and other companies.
Principle of HT-NGS involves the DNA molecules, which are sequenced in a massively parallel fashion in a flow cell (Mardis 2008a, b; Metzker 2010). The sequencing is conducted in either a stepwise iterative process or in a continuous real-time manner. By virtue of this highly parallel process, each clonal template or single molecule is “individually” sequenced and can be counted among the total sequences generated. The high-throughput combination of qualitative and quantitative sequence information generated has allowed advanced genome analyses that were previously, either not technically possible or cost prohibitive.
Second generation HT-NGS platforms
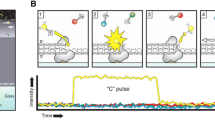
The second generation HT-NGS platforms can generate about five hundred million bases of raw sequence (Roche) to billions of bases in a single run (Illumina, SOLiD). These novel methods rely on parallel, cyclic interrogation of sequences from spatially separated clonal amplicons (26 μm oil-aqueous emulsion bead [Roche: pyrosequencing chemistry], 1 μm clonal bead [SOLiD: sequencing by sequential ligation of oligonucleotide probes], clonal bridge [Illumina: sequencing by reversible dye terminators]). Currently, these (above mentioned) three leading second generation HT-NGS platforms (Fig. 1) are commercially available and the race for more additional platforms are continuously on the horizon (for comprehensive reviews on complete laboratory methods, technical aspacts of sample preparation and resulting sequencing data analysis of Roche, Illumina, SOLiD platforms, see: Mardis 2008a, b, 2009, 2010; Metzker 2010). In 2008, the US National Human Genome Research Institute (NHGRI) has initiated funding for a series of projects as part of its revolutionary genome sequencing technologies program and aimed toward its target goal of sequencing a human genome for $1000 or less (http://www.genome.gov/27527585). Recently in December 2010, the NHGRI consortium has published the most comprehensive map of human genetic variation using next-generation DNA sequencing technologies to systematically characterize the genetic differences among 179 individuals from four populations and 697 individuals from seven populations in three pilot studies (Durbin et al. 2010). These pilot studies of the “1000 genomes project” laid a critical foundation for studying human genetic variation, and aimed to create a comprehensive, publicly available map of genetic variation, that will ultimately collect sequence from 2,500 people from multiple populations worldwide and underpin future genetics research (http://www.genome.gov/27541917).
Third generation HT-NGS platforms
In the previously discussed second generation HT-NGS platforms, the principle was based on the emulsion PCR amplification of DNA fragments, to make the light signal strong enough for reliable base detection by the CCD cameras. Although the PCR amplification has revolutionized DNA analysis, but in some instances it may introduce base sequence errors or favor of certain sequences over others, thus changing the relative frequency and abundance of various DNA fragments that existed before amplification. To overcome this, the ultimate miniaturization into the nanoscale and the minimal use of biochemicals, would be achievable if the sequence could be determined directly from a single DNA molecule, without the need for PCR amplification and its potential for distortion of abundance levels. This sequencing from a single DNA molecule is now called as the “third generation of HT-NGS technology” (Schadt et al. 2010). The concept of sequencing-by-synthesis without a prior amplification step, i.e., single-molecule sequencing is currently pursued by a number of companies and described below in Sects. 5.1 to 5.7.
Heliscope™ single molecule sequencer
One of the first techniques for sequencing from a single DNA molecule was introduced by Braslavsky et al. 2003 and licensed by Helicos biosciences as the first commercial single-molecule DNA sequencing system in 2007. The principle of Heliscope sequencer relies on “true single molecule sequencing” (tSMS) technology. The tSMS technology begins with DNA library preparation through DNA shearing and addition of poli-(A) tail to generated DNA fragments (Ozsolak et al. 2010), followed by hybridization of DNA fragments to the poli-(T) oligonucleotides which are attached to the flow cell and simultaneously sequenced in parallel reactions. The sequencing cycle consists of DNA extension with one, out of four fluorescently labeled nucleotides, followed by nucleotide detection with the Heliscope sequencer. The subsequent chemical cleavage of fluorophores allows the next cycle of DNA elongation to begin with another fluorescently labeled nucleotide, which enables the determination of the DNA sequence (Harris et al. 2008). The Heliscope sequencer is capable of sequencing up to 28 Gb in a single sequencing run and takes about 8 days. It can generate short reads with a maximal length of 55 bases. In a recent development, Helicos announced that it has developed a new generation of “one-base-at-a-time” nucleotides which allow more accurate homopolymer and direct RNA sequencing (Ozsolak and Milos 2011a, b).
Single molecule real time (SMRT™) sequencer
The principle of SMRT sequencer relies on single molecule real time sequencing by synthesis method provided on the sequencing chip containing thousands of zero-mode waveguides (ZMWs). The sequencing reaction of a DNA fragment is performed by a single DNA polymerase molecule, which is attached to the bottom of each ZMW so that each DNA polymerase resides at the detection zone of ZMW (Fig. 2).
During the sequencing reaction, the DNA fragment is elongated by DNA polymerase with dNTP’s that are fluorescently labeled (each nucleotide is labeled with a fluorophore of different color) at the terminal phosphate moiety. The DNA sequence is determined with CCD array on the basis of fluorescence nucleotide detection, which is performed before nucleotide incorporation, while the labeled dNTP forms a cognate association with the DNA template. The fluorescence pulse is stopped after phosphodiester bond formation, which causes the release of a fluorophore that diffuses out of ZMW. Subsequently, the labeled nucleotide incorporation and detection allow us to determine the DNA sequence (Levene et al. 2003; Eid et al. 2009). The SMRT sequencer was designed and is still being developed by the Pacific Biosciences (www.pacificbiosciences.com). Although the SMRT instrument has recently been available on the market, the company claims that the SMRT analyzer can be capable of obtaining 100 Gb per hour with reads longer than 1000 in a single run.
Single molecule real time (RNAP) sequencer
A different single-molecule DNA sequencing approach, i.e., RNA polymerase (RNAP), has been proposed by (Greenleaf and Block 2006) in which the RNAP is attached to one polystyrene bead, whilst the distal end of a DNA fragment is attached to another bead. Each bead is placed in an optical trap and the pair of optical traps levitated the beads. The RNAP interacts with the DNA fragment and the transcriptional motion of RNAP along the template, changes the length of the DNA between the two beads. This leads to displacement of the two beads that can be registered with precision in the Angstrom range, resulting in single-base resolution on a single DNA molecule. By aligning four displacement records, each with a lower concentration of one of the four nucleotides, in a role analogous to the primers used in Sanger sequencing and for calibration using the known sequences flanking to the unknown sequenced fragment, it is possible to deduce the sequence information. The technique demonstrates that the movement of a nucleic acid enzyme and the very sensitive optical trap method, which may allow extraction of sequence information directly from a single DNA molecule.
Nanopore DNA sequencer
In contrary to all DNA sequencers mentioned above, sequencing a DNA molecule with the Nanopore DNA sequencer is free of nucleotide labeling and detection. This technique was developed from studies on translocation of DNA through various artificial nanopores. The DNA sequencing with Nanopore instrument relies on the converting of electrical signal of nucleotides by passing through a nanopore which is an α-hemolysin pore covalently attached with cyclodextrin molecule – the binding site for nucleotides. The principle of this technique is based on the modulation of the ionic current through the pore as a DNA molecule traverses it, revealing characteristics and parameters (diameter, length and conformation) of the molecule (Fig. 2). During the sequencing process the ionic current that passes through the nanopore is blocked by the nucleotide, i.e., the previously cleaved by exonuclease from a DNA strand that interacts with cyclodextrin. The time period of current block is characteristic for each base and enables the DNA sequence to be determined (Astier et al. 2006; Rusk 2009). However, further improvements and modifications in the technique, for example, increasing the number of parameters measured during the translocation of the DNA enabling single-base resolution, could lead to a rapid nanopore-based DNA sequencing technique.
Real time single molecule DNA sequencer platforms developed by VisiGen Biotechnologies
The VisiGen biotechnologies (www.visigenbio.com) introduced a specially engineered DNA polymerase, which acting as a ‘real-time sensor’ for modified nucleotides with a donor fluorescent dye and incorporated close to the active site involved in selection of the nucleotides during synthesis (Fig. 2). All four nucleotides to be integrated were modified, each with a different acceptor dye. During the synthesis, when the correct nucleotide is found, it selected and entered into the active site of the enzyme, and the donor dye label in the polymerase came into close proximity with the acceptor dye on the nucleotides and energy was transferred from donor to acceptor dye giving rise to a fluorescent resonant energy transfer (FRET) light signal (Selvin 2000). The frequency of this signal varied depending on the label incorporated in the nucleotides, so that by recording frequencies of emitted FRET signals was possible to determine base sequences, at the speed at which the polymerase can integrate the nucleotides during the synthesis process (usually a few hundred per second). The acceptor fluorophore is removed during nucleotide incorporation, which ensures that there are no DNA modifications that might slow down the polymerase during synthesis. The company is currently working on its first version of the instrument, which can generate around 4 Gb of data per day. The single-molecule approach requires no cloning and no amplification, which eliminates a large part of the cost, relative to current technologies. In addition, read lengths for the instrument are expected to be around 1 kb, longer than any current platform.
Multiplex polony technology
Run by the privately-funded personal genome project (PGP) and lead by Prof. G Church’s research group (www.personalgenomes.org), has developed and introduced the multiplex polony technology (Mitra et al. 2003; Shendure et al. 2005). In this technique, several hundred sequencing templates are deposited onto thin agarose layers and sequences are determined in parallel. This metod presents increase of several orders of magnitudes in the number of samples which can be analyzed simultaneously. It has the advantage, in terms of large reduction of the reaction volumes, requiring smaller amounts of reagents and the resulting at a lower cost. The designed instrument, i.e., Danaher Motion Polonator model G.007, is capable of 10 to 35 Gbp per module per 2.5 day run. Instrument can couple with 200 of these modules to collect 100 diploid genomes at 30X coverage in 5 days, with the remaining 5 days used for repeating any weak runs to assure 98% coverage at 1E-5 accuracy. With the significant reduced volume of reagents, the cost per unit volume is lower about 10-fold and the company hopes to meet the goal of $1000 per genome soon.
The Ion Torrent sequencing technology
In a recent advancement, the first PostLightTM sequencing technology (Ion Torrent) has been introduced (http://www.iontorrent.com/). This technology creates a direct connection between the chemical and the digital information, enabling fast, simple, massively scalable sequencing. It utilizes the simple nucleic acid Watson’s chemistry to incredibly powerful, proprietary of semiconductor technology -The Moore’s law (Moore 1965). The principle of Ion Torrent semiconductor technology is based on a well-characterized biochemical process, in which a nucleotide is incorporated into a strand of DNA by a polymerase, resulting in a release of hydrogen ion as a byproduct (Fig. 2). The technological device uses a high-density array of micro-machined wells to perform this biochemical process in a massively parallel way with each well holding a different DNA template. Beneath the wells is an ion-sensitive layer and beneath that a proprietary Ion sensor. The massive parallel sequencing at Ion Personal Genome Machine (PGM™) sequencer works on the basis of “base” principle. For example, if nucleotide A is added to a DNA template and it is incorporated into a strand of DNA and then a hydrogen ion will be released. The charge from that ion will change the pH of the solution and can be detected directly by the ion sensor without scanning, cameras and light. In this way, the PGM™ sequencer sequentially floods the chip with one nucleotide after another. The designed PGM™ system enables to perform wide range of sequencing application such as, multiplexing amplicons, transcriptome, small RNA, and ChIP-Seq. paired-end sequencing and methylation.
In terms of issues related to genomics data quality and analysis, a substantial $10 million funding has been offered by the Archon Genomics X PRIZE (AGXP), in order to generate rapid, accurate and complete human DNA sequences to global research community (editorial discussion: Toward a medical grade human genome sequence. Nat Genet. 2011 Mar, 43 [3]: 173). Because so many genome researchers have a stake, AGXP offers to help with a process of community consultation to help evolve fair and efficient methods to validate contestant genome data at high degrees of accuracy and completeness (Kedes et al. 2011). Since the launch of AGXP in 2006, there have been important advances in validation protocols of DNA sequencing technologies both in terms of speed and reduction in costs (Sutton et al. 2011). However, no current human genome sequence is fully complete, fully accurate or certain to contain all rearrangements or information of chromosome phasing (haplotype). Highly repetitive and other genome-wide regions remain difficult to sequence but are likely to be critical in defining heritable features. Hence the ideals of the X Prize remain as critical for the future of human genetics and genetic medicine as ever.
Comparison of second and third HT-NGS platforms
Unlike the second HT-NGS technologies, that rely on PCR to grow clusters of a given DNA template, attaching the clusters of DNA templates to a solid surface that is subsequently imaged as the clusters are sequenced by synthesis in a phased approach, the third HT-NGS technologies interrogate single molecules of DNA in a such a way that no synchronization (a limitation of second HT-NGS) is required (Whiteford et al. 2009), thereby overcoming issues related to the biases introduced by PCR amplification and dephasing. Furthermore, third HT-NGS technologies have the potential to exploit more fully, the high catalytic rates and high processivity of DNA polymerase, or avoid any biology or chemistry altogether to radically increase read length (from tens of bases, to tens of thousands of bases per read) and time to result (from days, to hours, or minutes). Besides this, the third HT-NGS technologies may offer the following advantages over second HT-NGS technologies: i) higher throughput, ii) faster turnaround time (e.g., sequencing metazoan genomes at high fold coverage in minutes), iii) longer read lengths to enhance de novo assembly and enable direct detection of haplotypes and even whole chromosome phasing, iv) higher consensus accuracy to enable rare variant detection, v) small amounts of starting material (theoretically only a single molecule may be required for sequencing), and vi) low cost, where sequencing the human genome at high fold coverage for less than $1000 is now a reasonable goal for the community.
In the past six years, an influx of plenty of original as well as comprehensive review papers related to both second and third generation HT-NGS platforms have been published. Thereby, the comparison of second HT-NGS platforms (Roche/454, SOLiD, and Illumina) and third HT-NGS platforms (Helicos and Pacific Biosciences etc.) are summarized in Table 1, illustrating the similarities and differences in these technologies, according to several metrics. For examples, in terms of technological features, both platforms work sequencing by synthesis, however, Second HT-NGS platform deals with washing and scan of many copies of the DNA molecules in comparison to direct physical inspection of the DNA molecule and its resolution in real time (i.e., no protracted cycles of hybridization or successive enzymatic steps) in third HT-NGS platforms. Among other dissimarities are the RNA sequencing, where, second HT-NGS platform only performed the cDNA sequencing whereas direct RNA sequencing in case of third HT-NGS platforms. Regarding data analysis, both platforms have complexity because of large data volume. In second HT-NGS platforms the mojor challenges are the short reads which can be complicated in genome assembly and alignment algorithms, whereas new signal processing challenges are still prominent in the third HT-NGS platforms.
With the progressive advent of HT-NGS technologies, DNA sequencing costs have been drastically reduced (Table 1). Now, it is feasible to sequence hundreds or even thousands of genes for a single individual with a suspected genetic disease or complex disease predisposition. Along with the benefits offered by these technologies, there are a number of challenges that must be addressed before wide-scale sequencing becomes accepted in genome research practices. Molecular diagnosticians will need to become comfortable with, and gain confidence with, these new platforms, which are based on radically different technologies compared to the standard DNA sequencers in routine diagnostic today. Since 2001, when the technology that sequenced the human genome on the basis of capillary electrophoresis of individual fluorescently labeled Sanger sequencing method, the advent of next-generation sequencing platforms have dramatically increased the speed at which DNA sequence can be acquired, while reducing the costs by several orders of magnitude compared to their predecessors (Fig. 3). This is because of the basic mechanisms for data generation had changed radically, producing far more sequence reads per instrument run and at a significantly lower expense. Figure 3 illustrates how the resulting HT-NGS information has both enhanced our knowledge and expanded the impact of the genome on biomedical research (Mardis 2011).
Changes in instrument capacity over the past decade, and the timing of major sequencing projects (Figure updated from Madris, 2011). On top: increasing scale of data output per run plotted on a logarithmic scale. At middle: timeline representing major milestones in massively parallel sequencing platform introduction and instrument revisions. On bottom: the time lines of several projects, milestones and publications citing different sequencing technologies
These next-generation platforms generate shorter reads with lower quality, when compared to the Sanger platform. The reduction in read length and quality necessitated the development of bioinformatics tools to assist in either the mapping of these shorter reads to reference sequences or de novo assembly. The development of these new techniques aims toward meeting the demand for sequence information in various fields of research, such as study of genomics and evolution, forensics, epidemiology and diagnostics and applied therapeutics.
Applications and advances of sequencing technologies on human genome research
The landmark of sequencing of human genome was accomplished by two groups, i.e., the publicly funded Human Genome Group (HGP) and Celera Groups. Both groups utilized different strategies. The HGP group produced a working draft of the human genome by a map-based strategy, while Celera, to sequence the human genome by the whole-genome shotgun (WGS) approach (Fig. 4). The availability of sequence material obtained through different approaches greatly facilitated the ability of the entire scientific community to interpret the data. The strategy of HGP originally established by the publicly funded effort and was based on the localizing bacterial artificial chromosomes (BACs) containing large fragments of human DNA within the framework of a landmark-based physical map. Ideally, sequencing would have been done on a clone-by-clone basis, with clones selected from the minimum BAC tiling path. The key to the HGP's strategy was the subsequent 'mapping' step in which the BACs were each positioned on the genome's chromosomes by looking for distinctive marker sequences, called sequence tagged sites (STSs), whose location had already been pinpointed. In this way, the BACs provided a high-resolution map of the entire genome (Fig. 4). The working draft, although containing some gaps and ambiguities in order, is extremely useful in such efforts as identifying disease-associated genes. Simultaneously, the idealized strategy of Celera was to avoid the up-front mapping phase by subcloning random fragments of the human genome directly. Sequencing of both ends of fragments in libraries of different sizes facilitated ordering. While saving time and effort at the beginning, the Celera approach made the assembly process much more dependent on algorithms and computer time. In their efforts to reach their goals, the idealized strategies evolved into hybrids in which the HGP selected more clones arbitrarily and Celera made use of BAC maps and sequence generated by the HGP (Fig. 4).
Two strategies for generating the first complete human genome sequence: On right, the Human Genome Project (HGP) map-based strategy (Lander et al. 2001) showing the localizing bacterial artificial chromosomes (BACs) containing large fragments of human DNA within the framework of a landmark-based physical map. On left, the Celera’s strategy (Venter et al. 2001) showing the whole genome shotgun sequencing approach
Since the introduction of HT-NGS platform in 2005, the production of large numbers of low-cost reads made the NGS platforms useful for many applications on human genomes research particularly, the de novo genome sequencing, whole-genome resequencing or more targeted sequencing, cataloguing the transcriptomes of cells tissues and organisms (RNA–seq), genomic variation and mutation detection, genome-wide profiling of epigenetic marks and chromatin structure using methyl– seq, DNase–seq and ChIP–seq (chromatin immunoprecipitation coupled to DNA microarray) and personal genomics (Table 2).
De Novo, resequencing and targeted sequencing
In general, the HT-NGS platforms made de novo assembly of most organisms including human, a lengthy and costly endeavor. In humans, such an endeavor had already commenced with the publication of several complete genomes, for example: using the Roche 454 technology to 7.5x human genome coverage (Wheeler et al. 2008), human genome sequences of Chinese (Wang et al. 2008), an African (Pushkarev et al. 2009), and two Korean individuals (Ahn et al. 2009; Kim et al. 2009), all were done using the Illumina Genome Analyzer and sequenced around 20x haploid genome coverage with the exception of the African male’s genome which was also resequenced on ABI SOLiD system (McKernan et al. 2009). More recently, James Lupski’s genome was sequenced to 30x base coverage using ABI’s SOLiD System (Lupski et al. 2010). Resequencing of human genome was not limited to the second generation platforms. Steven Quake’s genome, for example, was sequenced to 90% genome coverage on Helicos’ single-molecule sequencing platform (Pushkarev et al. 2009). The whole genome genotyping approach on HT-NGS effectively enables unlimited multiplexing and unconstrained single nucleotide polymorphism (SNP) selection, for example typing of HLA genotypes in human (Lind et al. 2010) and genome-wide fetal genotyping using non-invasive HT-NGS of the mother's blood (Burgess 2011).
RNA sequencing
HT-NGS is also finding application in the study of small RNAs. For example, a comprehensive study of miRNA in acute myeloid leukaemia performed by HT-NGS identified differentially expressed miRNAs binding sites for acute myeloid leukaemia (Ramsingh et al. 2010). In recent studies, several efficient procedures have been introduced to perform RNA-Seq using the Illumina sequencing platform (Buermans et al. 2010; Nagalakshmi et al. 2010) including the technical issues (Marguerat and Bähler 2010), Construction of complex miRNA repertoire database (Lee et al. 2010), preparation the small RNA libraries and analyses of the resultant sequence data for measuring microRNA abundance (Morin et al. 2010), as well as annotation and discovery of small RNAs from transcriptomic data (Yang et al. 2011). RNA seq using Illumina and 454 technologies has also been found to be a powerful tool for detecting novel gene fusions in cancer cell lines and tissues (Maher et al. 2009). Understanding the transcriptome is essential for interpreting the functional elements of the genome and revealing the molecular constituents of cells and tissues, and also for understanding development and disease. The specific aims of transcriptomics are: (1) to catalog all transcripts in a context of cell types for a species, including mRNAs, non-coding RNAs and small RNAs, (2) to determine the transcriptional structure of genes, in terms of their start sites, 5’- and 3’-ends, splicing patterns and other post-transcriptional modifications and (3) to quantify the expression levels of each transcript during development or under different physiologic and pathological conditions. With the availability of faster and cheaper HT-NGS platforms, more transcriptomic analyses are performed using a recently-developed deep sequencing approach (Wang et al. 2009). The short reads produced by HT-NGS technologies, particularly Illumina and SOLiD, are arguably suitable for gene expression profiling. RNA-Seq has been used to accurately monitor gene expression of specific genes to determine the differential splicing, allele-specific expression of transcripts and many biological-related issues utilized in RNA-Seq experiments (Costa et al. 2010b).
Epigenetics
The HT-NGS technologies offer the potential to substantially accelerate epigenomic research (the study of heritable gene regulation that does not involve the DNA sequence itself but its modifications and higher-order structures), including posttranslational modifications of histones, the interaction between transcription factors and their direct targets, nucleosome positioning on a genome-wide scale and the characterization of DNA methylation patterns (Bormann et al. 2010; Fouse et al. 2010; Bhaijee et al. 2011). Histone modification and methylation of DNA are two important epigenetic mechanisms that regulate the transcriptional status of genes. Using ChIP-Seq (chromatin immunoprecipitation and direct sequencing) technology, post-translational modifications of histones and the location of transcription factors can be studied at the whole-genome level (Neff and Armstrong 2009), whereas methylated DNA immunoprecipitation (meDIP) and bisulphite protocols can be used to study the methylation of DNA itself (Popp et al. 2010). For example, using ChIP-seq on HT-NGS platform, the binding sites for a transcription factor (TF) and the human growth-associated binding protein (GABP alpha) were directly sequenced instead of being hybridized on a chip-array and unraveling the wide and intricate gene pathways regulated by PPARG gene (Costa et al. 2010a) and predicted the de novo motif discovery (Jiao et al. 2010). This ChIP-Seq on HT-NGS platform allows now researchers to improve both quantity and quality of produced data. Among other prevalent high-throughput approaches, protein-DNA interactions have been studied by the combination of chromatin immunoprecipitation with DNA microarray (ChIP-chip). Contrarily, ChIP-seq technique inherits two advantages from the HT-NGs platforms, firstly, it is not limited by the microarray content and secondly, it does not depend on the efficiency of probe hybridization. The ChIP-seq approach was recently used to identify binding sites of two transcription factors, STAT1 and NRSF in human cells (Robertson et al. 2007; Euskirchen et al. 2007). Both studies compared their findings with those generated by ChIP-chip, demonstrating that ChIP-seq had better resolution and required fewer replicates.
Genomic variation and mutation detection
NGS promises to facilitate the genome-wide human population structural variation studies (Xi et al. 2010; Henn et al. 2010) by uncovering all of the common and rare genetic variation in human populations (Bowne et al. 2011). Indeed, the “1000 Genomes Project” has made great progress to date toward this goal (Durbin et al. 2010). With a comprehensive genetic map of all human variation produced by NGS, researchers will be able to perform more detailed experiments to detect genetic variation underlying the response to medicines. HT-NGS patforms have also found application in high throughput mutation detection and carrier screening using a method called functional genomic fingerprinting (FGF). The method implies a selective enrichment of functional genomic regions (the exome, promoterome, or exon splice enhancers) approach in response to discovery of causal mutations for disease and drug response (Senapathy et al. 2010). The target enrichment based on microarray also allowed the parallel, large-scale analysis of complete genomic regions for multiple genes of a disease pathway, and for multiple samples simultaneously, thus providing an efficient tool for comprehensive diagnostic screening of mutations (Amstutz et al. 2011). The carrier screening by HT-NGS is also feasible to the general population with severe recessive childhood disorders (Bell et al. 2011) and in mutation detection associated with autosomal-recessive cerebellar ataxia, by combining SNP array-based linkage analysis and targeted resequencing of relevant sequences in the linkage interval (Vermeer et al. 2010).
Cancer research and biomarkers
HT- NGS technological advances are also driving the development of novel diagnostic and therapeutic approaches to the treatment of cancer (Meyerson et al. 2010), as researchers re-sequence the tumor and normal genomes comparision from specific types of cancer (Pfeifer and Hainaut 2011). In cancer genome, it enables rapid identification of patient-specific rearrangements in solid tumours (Ding et al. 2010, McBride et al. 2010). A further interesting application of the technology is ‘personalized’ biomarkers, which has recently been developed to detect the presence of tumour-specific genomic rearrangements in plasma samples from patients to generate a tumour-specific biomarker. Genomic rearrangements were identified specific to the tumour, which were not present in the patient's normal somatic tissue. Digital PCR assays were then designed across rearrangement breakpoints to provide a sensitive tumour-specific biomarker which was successfully used to monitor residual disease following treatment (Leary et al. 2010). Furthermore, HT-NGS technology also enables to identify the causal mutations responsible for driving cancer initiation and metastasis and raises significant expectations for improving oncologic outcomes (Katsios et al. 2010) and identification of 4883 SOX2 binding regions in the Glioblastoma (GBM) cancer genome (Fang et al. 2011). Small RNAs could provide a further application of NGS in biomarker discovery (Lee et al. 2010). MicroRNAs (miRNAs) are implicated in the control of protein translation and are present in blood plasma. NGS could be used to assay tissues or blood plasma to generate whole-genome miRNA profiles, which could then be mined for biomarker signatures.
Applications and advances of sequencing technologies on animal genome research
Discussion pertaining to the economically important farm animals in context to impact of HT-NGS technology is also a scope of this publication. The updated assembled animal genome server is available for the following species: cat (Felis catus), chicken (Gallus gallus), cow (Bos taurus), dog (Canis lupus familiaris), horse (Equus caballus) and pig (Sus scrofa) (http://www.ensembl.org/info/about/species.html). Additionally, the preview of genome assemble is also available for the sheep (Ovis aries) (http://pre.ensembl.org/Ovis_aries/Info/Index) and turkey (Meleagris gallopavo) genome (http://pre.ensembl.org/Meleagris_gallopavo/Info/Index). The important characteristics features of these assembled animal genomes are summarized in Table 3.
The first initiative toward animal genome sequencing started, just after the human genome sequencing conducted in 2001 (Lander et al. 2001; Venter et al. 2001) to advocate public funding for animal genomics research from several universities, private industries, producer groups and Animal Genome Research (AGR) scientific group. At the beginning, AGR under the National Academy of Sciences (NAS) organized a public workshop on “exploring horizons for domestic animal genomics” (Pool and Waddell 2002). The workshop objective was to identify research goals, public and private fundings to generate high-coverage, draft genome sequences of the major domestic animal species, i.e., cattle, pig, horse, sheep, chicken, dog and cat. In the initial phase, two “white papers” were released in support for sequencing and assembling the cattle genome (Gibbs et al. 2002) and pig genome (Rohrer et al. 2002). Since then, considerable progress has been achieved toward the whole genome sequence of domestic animals. In 2006, latest progress and updates on bovine (Womack 2006), porcine (Mote and Rothschild 2006), sheep (Cockett 2006), horse (Chowdhary and Raudsepp 2006), chicken (Burt 2006), canine (Galibert and André 2006) and feline (Murphy 2006) genome sequencing were comprehensively reviewed and published. For the recent progress and development in domestic animals genome sequencing and assembling, a series of publications and studies were published(Table 3), for bovine (Liu 2009; Elsik et al. 2009; Zimin et al. 2009), porcine (Wiedmann et al. 2008; Amaral et al. 2009; Ramos et al. 2009; Isom et al. 2010; Leifer et al. 2010), sheep (Archibald et al. 2010), horse (Bright et al. 2009; Coleman et al. 2010), chicken (Dalloul et al. 2010, Marklund and Carlborg 2010).
In the past, animal genome researches have effectively adapted to traditional breeding program objectives, i.e., genetic improvement of the livestock through marker assisted selection (MAS) and gene assisted selection (GAS) program. The MAS programs were successfully implemented during mid 1990s through global animal genome scan procedures and quantitative trait loci (QTL) mapping (for example cattle: Georges et al. 1995 and pig: Andersson et al. 1994). The GAS program was well implemented to animal genomes with identification of candidate genes and detection of casual quantitative trait nucleotides (QTN) discoveries (for example cattle: Grisart et al. 2001 and pig: Van Laere et al. 2003). In the past five years, HT-NGS technologies emerged as a potential research tool for the development of animal genomes research. Now, in this genomics era, the livestock researchers are working on effective implementation of genomics breeding for selection (GBS) in their traditional breeding program toward genetic improvement of economic traits.
In contrast to human, the progress to adopt the HT-NGS technologies in animal genome is at a lower pace, predominantly because of high cost and limited publically funded projects. However, continuous advancement of HT-NGS through the fast track human research and feasibility of genome sequencing for 1000 $, the pace will be geared up faster in the near future. The overall review on impact of current HT-NGS in animal genome is presented in Table 4.
Concluding summary and future prospective
In the fast growing HT-NGS technologies, the main challenge is to cope with the analysis of vast production of sequencing database through advanced bioinformatics tools. As the year 2011 has been marked as the 10th anniversary of the first human genome sequencing, nucleic acid research (NAR) has recently published its 18th annual database issue (Volume 39, supplement 1 January 2011: http://nar.oxfordjournals.org/content/39/suppl_1) to dedicate 10 years achievements in genome sequencing and its future challenges ahead. The published issues comprised of 96 new online databases covering a variety of molecular biology data and 83 data resources that have previously been published in NAR or other journals, in total the database collection now includes 1330 data sources.
The availability of ultra-deep sequencing of genomic DNA will transform the medical (in analysis of the causes of disease, development of new drugs and diagnostics) and veterinary (genetic improvement of animal health and productivity) fields in the near future. Further, it may become a promising tool in the analysis of chromatin immunoprecipitation coupled to DNA microarray (ChIP-chip) or sequencing (ChIP-seq), RNA sequencing (RNA-seq), whole genome genotyping, de novo assembling and re-assembling of genome, genome wide structural variation, mutation detection and carrier screening, detection of inherited disorders and complex human diseases, DNA library preparation, paired ends and genomic captures, sequencing of mitochondrial genome and personal genomics etc. It is anticipated that HT-NGS technology, for clinical purposes in human medicine and implementation of genomic selection in farm animals breeding programs will probably be fully adopted in the next couple of decades. The recent technological advancements in HT-NGS analysis is not only setting the benchmark in the advancement in genomics researches but also in the fields of proteomics, other omics and cancer research at a unprecedented pace (e.g., DNA and protein microarrays, quantitative PCR, mass spectrometry and others). Although, the HT-NGS with short DNA sequence reads (25–50 bases) and moderate sequence reads (500 bases) have already found many potential applications, but for genomic sequencing and for analysis of the ever more important structural genetic variations in genomes, such as copy number variations, chromosomal translocations, inversions, large deletions, insertions and duplications, it would be a great advantage if sequence read length on the original single DNA molecule could be increased to several 1000 bases and more per second. With the progress of third NGS platform at tremendous pace, one can hope that the goal of determining a whole chromosome sequence from a single original DNA molecule or genome sequence for $1000 could be feasible soon in the near future.
References
Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS et al. (2009) The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res 19:1622–1629
Amaral AJ, Megens HJ, Kerstens HH, Heuven HC, Dibbits B, Crooijmans RP, den Dunnen JT, Groenen MA (2009) Application of massive parallel sequencing to whole genome SNP discovery in the porcine genome. BMC Genomics 10:374
Amstutz U, Andrey-Zürcher G, Suciu D, Jaggi R, Häberle J, Largiadèr CR (2011) Sequence capture and next-generation resequencing of multiple tagged nucleic acid samples for mutation screening of urea cycle disorders. Clin Chem 57:102–111
Andersson L, Haley CS, Ellegren H, Knott SA et al. (1994) Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 263:1771–1774
Ansorge W et al. (1986) A non-radioactive automated method for DNA sequence determination. J Biochem Biophys Methods 13:315–323
Ansorge W et al. (1987) Automated DNA sequencing: ultrasensitive detection of fluorescent bands during electrophoresis. Nucleic Acids Res 15:4593–4602
Archibald AL, Cockett NE, Dalrymple BP, International Sheep Genomics Consortium et al. (2010) The sheep genome reference sequence: a work in progress. Anim Genet 41:449–453. doi:10.1111/j.1365-2052.2010.02100.x
Astier Y, Braha O, Bayley H (2006) Toward Single Molecule DNA Sequencing: Direct Identification of Ribonucleoside and Deoxyribonucleoside 5’-Monophosphates by Using an Engineered Protein Nanopore Equipped with a Molecular Adapter. J Am Chem Soc 128:1705–1710
Bansal V (2010) A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics 26:i318–i324
Bashamboo A, Ledig S, Wieacker P, Achermann JC, McElreavey K (2010) New technologies for the identification of novel genetic markers of disorders of sex development (DSD). Sex Dev 4:213–224
Bell CJ, Dinwiddie DL, Miller NA, Hateley SL et al. (2011) Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 3:65ra4
Bhaijee F, Pepper DJ, Pitman KT, Bell D (2011) New developments in the molecular pathogenesis of head and neck tumors: a review of tumor-specific fusion oncogenes in mucoepidermoid carcinoma, adenoid cystic carcinoma, and NUT midline carcinoma. Ann Diagn Pathol 15:69–77
Bormann G, Chung CA, Boyd VL, McKernan KJ, Fu Y, Monighetti C, Peckham HE, Barker M (2010) Whole methylome analysis by ultra-deep sequencing using two-base encoding. PLoS One 5:e9320
Bowne SJ, Sullivan LS, Koboldt DC, Ding L et al. (2011) Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Invest Ophthalmol Vis Sci 52:494–503
Braslavsky I, Hebert B, Kartalov E, Quake SR (2003) Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci USA 100:3960–3964
Bright LA, Burgess SC, Chowdhary B, Swiderski CE, McCarthy FM (2009) Structural and functional-annotation of an equine whole genome oligoarray. BMC Bioinforma 11:S8
Buermans HP, Ariyurek Y, van Ommen G et al. (2010) New methods for next generation sequencing based microRNA expression profiling. BMC Genomics 11:716
Burgess DJ (2011) Human Disease: Next-generation sequencing of the next generation. Nat Rev Genet 12:78–79
Burt D, (2006) The Chicken Genome. In: Vertebrate Genomes, Volff J-N (ed). Genome Dyn. Karger 2:123–137
Chowdhary B, Raudsepp T (2006) The Horse Genome. In: Vertebrate Genomes, Volff J-N (ed). Genome Dyn. Karger 2:97–110
Cockett N (2006) The Sheep Genome. In: Vertebrate Genomes, Volff J-N (ed). Genome Dyn. Karger, 2:79–85
Coleman SJ, Zeng Z, Wang K, Luo S, Khrebtukova I, Mienaltowski MJ, Schroth GP, Liu J, MacLeod JN (2010) Structural annotation of equine protein-coding genes determined by mRNA sequencing. Anim Genet 41:121–130
Collins FS, Green ED, Guttmacher AE, Guyer MS (2003) A vision for the future of genomics research. Nature 422:835–847
Costa V, Gallo MA, Letizia F, Aprile M, Casamassimi A, Ciccodicola A (2010a) PPARG: Gene Expression Regulation and Next-Generation Sequencing for Unsolved Issues. PPAR Res 2010:409168
Costa V, Angelini C, De Feis I, Ciccodicola A (2010b) Uncovering the complexity of transcriptomes with RNA-Seq. J Biomed Biotechnol 2010:853916
Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K et al. (2010) Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol 8:e1000475
Dames S, Durtschi J, Geiersbach K, Stephens J, Voelkerding KV (2010) Comparison of the Illumina Genome Analyzer and Roche 454 GS FLX for resequencing of hypertrophic cardiomyopathy-associated genes. J Biomol Tech 21:73–80
Day-Williams AG, Zeggini E (2010) The effect of next-generation sequencing technology on complex trait research. Eur J Clin Invest. doi:10.1111/j.1365-2362.2010.02437.x
Ding L, Wendl MC, Koboldt DC, Mardis ER (2010) Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet 19:R188–R196
Drögemüller C, Tetens J, Sigurdsson S, Gentile A, Testoni S, Lindblad-Toh K, Leeb T (2010) Identification of the bovine Arachnomelia mutation by massively parallel sequencing implicates sulfite oxidase (SUOX) in bone development. PLoS Genet 6:e1001079
Durbin RM, Abecasis GR, Altshuler DL, The 1000 Genomes Project Consortium et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073
Eck SH, Benet-Pagès A, Flisikowski K, Meitinger T, Fries R, Strom TM (2009) Whole genome sequencing of a single Bos taurus animal for single nucleotide polymorphism discovery. Genome Biol 10:R82
Edwards A et al. (1990) Automated DNA sequencing of the human HPRT locus. Genomics 6:593–608
Edwards CJ, Magee DA, Park SD, McGettigan PA et al. (2010) A complete mitochondrial genome sequence from a mesolithic wild aurochs (Bos primigenius). PLoS One 5:e9255
Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G et al. (2009) Real-time DNA sequencing from single polymerase molecules. Science 323:133–138
Elsik CG, Tellam RL et al. (2009) The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324:522–528
Erhard F, Zimmer R (2010) Classification of ncRNAs using position and size information in deep sequencing data. Bioinformatics 26:i426–i432
Euskirchen GM, Rozowsky JS, Wei CL et al. (2007) Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res 17:898–909
Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B (2011) The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics 12:11
Farias-Hesson E, Erikson J, Atkins A, Shen P, Davis RW, Scharfe C, Pourmand N (2010) Semi-automated library preparation for high-throughput DNA sequencing platforms. J Biomed Biotechnol 2010:617469
Fouse SD, Nagarajan RP, Costello JF (2010) Genome-scale DNA methylation analysis. Epigenomics 2:105–117
Galibert F, André C, (2006) The Dog Genome. In: Vertebrate Genomes, Volff J-N (ed). Genome Dyn. Karger, 2:46–59
Georges M, Nielsen D, Mackinnon M, Mishra A, Okimoto R et al. (1995) Mapping Quantitative Trait Loci Controlling Milk Production in Dairy Cattle by Exploiting Progeny Testing. Genetics 139:907–920
Gibbs R, Weinstock G, Rohrer G, Kappes S, Schook L, Skow L, Womack J (2002) Bovine genomic sequencing initiative Cattle-izing the human genome. Bovine sequencing white paper. pp: 1–12. http://www.genome.gov/pages/research/sequencing/seqproposals/bovineseq.pdf
Green RD, Qureshi MA, Long JA, Burfening PJ, Hamernik DL (2007) Identifying the Future Needs for Long-Term USDA Efforts in Agricultural Animal Genomics. Int J Biol Sci 3:185–191
Greenleaf WJ, Block SM (2006) Single-molecule, motion-based DNA sequencing using RNA polymerase. Science 313:801
Grisart B, Coppieters W, Farnir F, Karim L, Ford C et al. (2001) Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res 12:222–231
Hackenberg M, Barturen G, Oliver JL, Smeth NG (2011) A database for next-generation sequencing single-cytosine-resolution DNA methylation data. Nucleic Acids Res 39:D75–D79
Hajirasouliha I, Hormozdiari F, Alkan C, Kidd JM, Birol I, Eichler EE, Sahinalp SC (2010) Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics 26:1277–1283
Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M et al. (2008) Single-molecule DNA sequencing of a viral genome. Science 320:106–109
Henn BM, Gravel S, Moreno-Estrada A, Acevedo-Acevedo S, Bustamante CD (2010) Fine-scale population structure and the era of next-generation sequencing. Hum Mol Genet 19:R221–R226
Hestand MS, Klingenhoff A, Scherf M, Ariyurek Y et al. (2010) Tissue-specific transcript annotation and expression profiling with complementary next-generation sequencing technologies. Nucleic Acids Res 38:e165
Hillier LW, Miller W, Birney E, Warren W, International Chicken Genome Sequencing Consortium et al. (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Hong D, Park SS, Ju YS, Kim S, Shin JY et al. (2011) TIARA: a database for accurate analysis of multiple personal genomes based on cross-technology. Nucleic Acids Res 39:D883–D888
Isom SC, Spollen WG, Blake SM, Bauer BK, Springer GK, Prather RS (2010) Transcriptional profiling of day 12 porcine embryonic disc and trophectoderm samples using ultra-deep sequencing technologies. Mol Reprod Dev 77:812–819
Jex AR, Hall RS, Littlewood DT, Gasser RB (2010) An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Res 38:522–533
Jiao S, Bailey CP, Zhang S, Ladunga I (2010) Probabilistic peak calling and controlling false discovery rate estimations in transcription factor binding site mapping from ChIP-seq. Methods Mol Biol 674:161–177
Katsios C, Ziogas DE, Liakakos T, Zoras O, Roukos DH (2010) Translating Cancer Genomes Sequencing Revolution into Surgical Oncology Practice. J Surg Res. doi:10.1016/j.jss.2010.10.038
Kedes L, Liu E, Jongeneel CV, Sutton G (2011) Judging the Archon Genomics X PRIZE for whole human genome sequencing. Nature Genet 43:175
Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA et al. (2009) A highly annotated whole-genome sequence of a Korean individual. Nature 460:1011–1015
Kingsley CB (2011) Identification of causal sequence variants of disease in the next generation sequencing era. Methods Mol Biol 700:37–46
Kuhlenbäumer G, Hullmann J, Appenzellerm S (2011) Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat 32:144–151
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K et al. (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C et al. (2010) Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2:20ra14
Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K (2010) Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 16:2170–2180
Leifer I, Hoffmann B, Höper D, Bruun Rasmussen T, Blome S, Strebelow G, Höreth-Böntgen D, Staubach C, Beer M (2010) Molecular epidemiology of current classical swine fever virus isolates of wild boar in Germany. J Gen Virol 91:2687–2697
Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW (2003) Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299:682–686
Li R, Zhu H, Ruan J, Qian W, Fang X et al. (2010a) de novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Li R, Fan W, Tian G, Zhu H, He L, Cai J et al. (2010b) The sequence and de novo assembly of the giant panda genome. Nature 463:311–317
Lind C, Ferriola D, Mackiewicz K, Heron S, Rogers M et al. (2010) Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Hum Immunol 71:1033–1042
Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB et al. (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819
Liu GE (2009) Applications and case studies of the next-generation sequencing technologies in food, nutrition and agriculture. Recent Pat Food Nutr Agric 1:75–79
Liu Y, Qin X, Henry Song X et al. (2009) Bos taurus genome assembly. BMC Genomics 10:180
Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC et al. (2010) Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med 362:1181–1191
MacHugh DE, Gormley E, Park SD, Browne JA, Taraktsoglou M, O'Farrelly C, Meade KG (2009) Gene expression profiling of the host response to Mycobacterium bovis infection in cattle. Transbound Emerg Dis 56:204–214
Mardis ER (2008a) Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9:387–402
Mardis ER (2008b) The impact of next-generation sequencing technology on genetics. Trends Genet 3:133–141
Mardis ER (2009) New strategies and emerging technologies for massively parallel sequencing: applications in medical research. Genome Med 1:40
Mardis ER (2010) The $1,000 genome, the $100,000 analysis? Genome Med 2:84
Mardis ER (2011) A dacade’s perspective on DNA sequencing technology. Nature 470:198–203
Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S et al. (2009) Transcriptome sequencing to detect gene fusions in cancer. Nature 458:97–101
Marguerat S, Bähler J (2010) RNA-seq: from technology to biology. Cell Mol Life Sci 67:569–579
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Marklund S, Carlborg O (2010) SNP detection and prediction of variability between chicken lines using genome resequencing of DNA pools. BMC Genomics 11:665
Maxam AM, Gilbert W (1977) A new method for sequencing DNA. Proc Natl Acad Sci USA 74:560–564
McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ et al. (2010) Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer 49:1062–1069
McComish BJ, Hills SF, Biggs PJ, Penny D (2010) Index-free de novo assembly and deconvolution of mixed mitochondrial genomes. Genome Biol Evol 2:410–424
McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y et al. (2009) Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res 19:1527–1541
Metzker ML (2010) Sequencing technologies- the next generation. Nat Rev Genet 1:31–46
Meyerson M, Gabriel S, Getz G (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11:685–696
Michelizzi VN, Dodson MV, Pan Z, Amaral ME, Michal JJ, McLean DJ, Womack JE, Jiang Z (2010) Water buffalo genome science comes of age. Int J Biol Sci 6:333–349
Michelizzi VN, Wu X, Dodson MV, Michal JJ, Zambrano-Varon J, McLean DJ, Jiang Z (2011) A Global View of 54,001 Single Nucleotide Polymorphisms (SNPs) on the Illumina BovineSNP50 BeadChip and their transferability to Water Buffalo. Int J Biol Sci 7:18–27
Mitra RD et al. (2003) Digital genotyping and haplotyping with polymerase colonies. Proc Natl Acad Sci USA 100:5926–5931
Moore G (1965) Cramming more components onto integrated circuits. Electronics 38
Morin RD, Zhao Y, Prabhu AL, Dhalla N, McDonald H, Pandoh P, Tam A, Zeng T, Hirst M, Marra M (2010) Preparation and analysis of microRNA libraries using the Illumina massively parallel sequencing technology. Methods Mol Biol 650:173–199
Mote B, Rothschild M (2006) Cracking the genomic piggy bank: Identifying secrets of the pig genome. In: Vertebrate Genomes, Volff J-N (ed). Genome Dyn. Karger, 2:86–96
Murphy W (2006) The Feline Genome. In: Vertebrate Genomes, Volff JN (ed). Genome Dyn. Karger, 2:60–68
Nagalakshmi U, Waern K, Snyder M (2010) RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol Chapter 4(Unit 4.11):1–13
Nagarajan N, Pop M (2010) Sequencing and genome assembly using next-generation technologies. Methods Mol Biol 673:1–17
Neff T, Armstrong SA (2009) Chromatin maps, histone modifications and leukemia. Leukemia 23:1243–1251
Nyren et al. (1993) Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal Biochem 208:171–175
Nyren P (2007) The history of pyrosequencing. Methods Mol Biol 373:1–14
Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM (2010) Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143:1018–1029
Ozsolak F, Milos PM (2011a) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98
Ozsolak F, Milos PM (2011b) Transcriptome profiling using single-molecule direct RNA sequencing. Methods Mol Biol 733:51–61
Pérez-Enciso M, Ferretti L (2010) Massive parallel sequencing in animal genetics: wherefroms and wheretos. Anim Genet 41:561–569. doi:10.1111/j.1365-2052.2010.02057.x
Pfeifer GP, Hainaut P (2011) Next-generation sequencing: emerging lessons on the origins of human cancer. Curr Opin Oncol 23:62–68
Pontius JU, Mullikin JC, Smith DR, Lindblad-Tohet K, Team AS et al. (2007) Initial sequence and comparative analysis of the cat genome. Genome Res 17:1675–1689
Pool R, Waddell K (2002) Exploring horizons for domestic animal genomics. Workshop summary. National Academic Press, Constitution Avenue, NW, Washington, D.C. (www.nap.edu).
Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1105
Pushkarev D, Neff NF, Quake SR (2009) Single-molecule sequencing of an individual human genome. Nat Biotechnol 27:847–850
Ramos AM, Crooijmans RP, Affara NA, Amaral AJ, Archibald AL et al. (2009) Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4:e6524
Ramsingh G, Koboldt DC, Trissal M, Chiappinelli KB et al. (2010) Complete characterization of the microRNAome in a patient with acute myeloid leukemia. Blood 116:5316–5326
Raymond FL, Whittaker J, Jenkins L, Lench N, Chitty LS (2010) Molecular prenatal diagnosis: the impact of modern technologies. Prenat Diagn 30:674–681
Robertson G, Hirst M, Bainbridge M et al. (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4:651–657
Rohrer G, Beever JE, Rothschild MF, Schook L, Gibbs R, Weinstock G (2002) Porcine genomic sequencing initiative. Porcine sequencing white paper. pp: 1–10. http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/PorcineSEQ021203.pdf
Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P (1996) Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem 242:84–89
Ronaghi M, Uhlen M, Nyren P (1998) A sequencing method based on real-time pyrophosphate. Science 281:363–365
Rusk N (2009) Cheap third-generation sequencing. Nat Methods 6:244–245
Sanger F (1975) The Croonian Lecture, 1975: Nucleotide Sequences in DNA, B191 Proc. Royal Soc, London, 317
Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5467
Schadt EE, Turner S, Kasarskis A (2010) A window into third-generation sequencing. Hum Mol Genet 19:R227–R240
Schuster SC et al. (2008) Method of the year, next-generation DNA sequencing: Functional genomics and medical applications, Nat. Methods 5:11–21
Selvin PR (2000) The renaissance of fluorescence resonance energy transfer. Nat Struct Biol 7:730–734
Senapathy P, Bhasi A, Mattox J, Dhandapany PS, Sadayappan S (2010) Targeted genome-wide enrichment of functional regions. PLoS One 5:e11138
Shah AA, Meese E, Blin N (2010) Profiling of regulatory microRNA transcriptomes in various biological processes. a review. J Appl Genet 51:501–507
Shapiro B, Hofreiter M (2010) Analysis of ancient human genomes: using next generation sequencing, 20-fold coverage of the genome of a 4,000-year-old human from Greenland has been obtained. BioEssays 32:388–391
Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP et al. (2005) Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309:1728–1732
Singleton AB, Hardy J, Traynor BJ, Houlden H (2010) Towards a complete resolution of the genetic architecture of disease. Trends Genet 26:438–442
Smith LM et al. (1986) Fluorescence detection in automated DNA sequence analysis. Nature 321:674–679
Soroka M, Burzynski A (2010) Complete sequences of maternally inherited mitochondrial genomes in mussels Unio pictorum (Bivalvia, Unionidae). J Appl Genet 51:469–476
Sutton G, Liu E, and Jongeneel CV. Kedes L (2011) Archon genomics X PRIZE validation protocol. Nature Preceedings (http://precedings.nature.com/documents/5731/version/1).
Tawfik DS, Griffiths AD (1998) Man-made cell-like compartments for molecular evolution. Nature Biotech 16:652–656
Turner DJ, Keane TM, Sudbery I, Adams DJ (2009) Next-generation sequencing of vertebrate experimental organisms. Mamm Genome 20:327–338
Van Tassell CP, Smith TPL, Matukumalli LK, Taylor JF, Schnabel RD et al. (2008) SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat Methods 5:247–252
Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, Archibald AL, Haley CS, Buys N, Tally M, Andersson G, Georges M, Andersson L (2003) A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425:832–836
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO et al. (2001) The sequence of the human genome. Science 291:1304–1351
Vermeer S, Hoischen A, Meijer RP, Gilissen C, Neveling K et al. (2010) Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am J Hum Genet 87:813–819
Voelkerding KV, Dames S, Durtschi JD (2010) Next generation sequencing for clinical diagnostics-principles and application to targeted resequencing for hypertrophic cardiomyopathy: a paper from the 2009 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn 12:539–551
Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S et al. (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326:865–867
Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, Nord AS, Mandell JB, Swisher EM, King MC (2010) Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci USA 107:12629–12633
Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J et al. (2008) The diploid genome sequence of an Asian individual. Nature 456:60–65
Wang WC, Lin FM, Chang WC, Lin KY, Huang HD, Lin NS (2009) miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinforma 10:328
Werner T (2010) Next generation sequencing in functional genomics. Brief Bioinform 11:499–511
Wernersson R, Schierup MH, Jørgensen FG, Gorodkin J, Panitz F et al. (2005) Pigs in sequence space: A 0.66× coverage pig genome survey based on shotgun sequencing. BMC Genomics 6:70
Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A et al. (2008) The complete genome of an individual by massively parallel DNA sequencing. Nature 452:872–876
Whiteford N, Skelly T, Curtis C, Ritchie ME, Löhr A, Zaranek AW, Abnizova I, Brown C (2009) Swift: primary data analysis for the Illumina Solexa sequencing platform. Bioinformatics 25:2194–2199
Wiedmann RT, Smith TP, Nonneman DJ (2008) SNP discovery in swine by reduced representation and high throughput pyrosequencing. BMC Genet 9:81
Womack J (2006) The Bovine Genome. In: Vertebrate Genomes, Volff JN (ed). Genome Dyn. Karger 2:69–78
Wright CF, Morelli MJ, Thébaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP (2011) Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus using next-generation genome sequencing. J Virol 85:2266–2275
Xi R, Kim TM, Park PJ (2010) Detecting structural variations in the human genome using next generation sequencing. Brief Funct Genomics 9:405–415
Yamey G (2000) Scientists unveil first draft of human genome. BMJ 321:7
Yang JH, Shao P, Zhou H, Chen YQ, Qu LH (2011) deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res 38:D123–D130
Zimin AV, Delcher AL, Florea L, Kelley DR et al. (2009) A whole-genome assembly of the domestic cow. Bos Taurus Genome Biol 10:R42. doi:10.1186/gb-2009-10-4-r42
Zoghbi HY, Warren ST (2010) Neurogenetics: advancing the "next-generation" of brain research. Neuron 68:165–173
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Pareek, C.S., Smoczynski, R. & Tretyn, A. Sequencing technologies and genome sequencing. J Appl Genetics 52, 413–435 (2011). https://doi.org/10.1007/s13353-011-0057-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-011-0057-x