Abstract
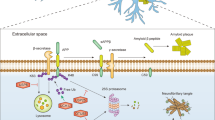
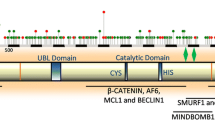
In healthy cell, inappropriate accumulation of poor or damaged proteins is prevented by cellular quality control system. Autophagy and ubiquitin proteasome system (UPS) provides regular cytoprotection against proteotoxicity induced by abnormal or disruptive proteins. E3 ubiquitin ligases are crucial components in this defense mechanism. Mahogunin Ring Finger-1 (MGRN1), an E3 ubiquitin ligase of the Really Interesting New Gene (RING) finger family, plays a pivotal role in many biological and cellular mechanisms. Previous findings indicate that lack of functions of MGRN1 can cause spongiform neurodegeneration, congenital heart defects, abnormal left-right patterning, and mitochondrial dysfunctions in mice brains. However, the detailed molecular pathomechanism of MGRN1 in cellular functions and diseases is not well known. This article comprehensively represents the molecular nature, characterization, and functions of MGRN1; we also summarize possible beneficiary aspects of this novel E3 ubiquitin ligase. Here, we review recent literature on the role of MGRN1 in the neuro-pathobiological mechanisms, with precise focus on the processes of neurodegeneration, and thereby propose new lines of potential targets for therapeutic intervention.




Similar content being viewed by others
References
Dobson CM (2003) Protein folding and misfolding. Nature 426(6968):884–890. doi:10.1038/nature02261
Duncan R, Hershey JW (1983) Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J Biol Chem 258(11):7228–7235
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147(4):789–802. doi:10.1016/j.cell.2011.10.002
Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G et al (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416(6880):507–511. doi:10.1038/416507a
Chen B, Retzlaff M, Roos T, Frydman J (2011) Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 3(8):a004374. doi:10.1101/cshperspect.a004374
Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355. doi:10.1146/annurev-biochem-060208-092442
Fimia GM, Kroemer G, Piacentini M (2013) Molecular mechanisms of selective autophagy. Cell Death Differ 20(1):1–2. doi:10.1038/cdd.2012.97
Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Furst DO, Saftig P et al (2010) Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 20(2):143–148. doi:10.1016/j.cub.2009.11.022
Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, Vakeel P, Stadel D et al (2013) Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol 23(5):430–435. doi:10.1016/j.cub.2013.01.064
Kaushik S, Cuervo AM (2012) Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 22(8):407–417. doi:10.1016/j.tcb.2012.05.006
Kettern N, Dreiseidler M, Tawo R, Hohfeld J (2010) Chaperone-assisted degradation: multiple paths to destruction. Biol Chem 391(5):481–489. doi:10.1515/BC.2010.058
Green M (1989) Genetic variants and strains of the laboratory mouse., vol 12–403. Catalog of mutant genes and polymorphic loci. Oxford University Press, New York
Lane PW, Green MC (1960) Mahogany, a recessive color mutation in linkage group v of the mouse. J Hered 51(5):228–230
Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1998) Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res 5(1):31–39
Phan LK, Lin F, LeDuc CA, Chung WK, Leibel RL (2002) The mouse mahoganoid coat color mutation disrupts a novel C3HC4 RING domain protein. J Clin Invest 110(10):1449–1459. doi:10.1172/JCI16131
He L, Lu XY, Jolly AF, Eldridge AG, Watson SJ, Jackson PK, Barsh GS, Gunn TM (2003) Spongiform degeneration in mahoganoid mutant mice. Science 299(5607):710–712. doi:10.1126/science.1079694
He L, Eldridge AG, Jackson PK, Gunn TM, Barsh GS (2003) Accessory proteins for melanocortin signaling: attractin and mahogunin. Ann N Y Acad Sci 994:288–298
Bagher P, Jiao J, Owen Smith C, Cota CD, Gunn TM (2006) Characterization of mahogunin ring finger-1 expression in mice. Pigment Cell Res 19(6):635–643. doi:10.1111/j.1600-0749.2006.00340.x
Perez-Oliva AB, Olivares C, Jimenez-Cervantes C, Garcia-Borron JC (2009) Mahogunin ring finger-1 (MGRN1) E3 ubiquitin ligase inhibits signaling from melanocortin receptor by competition with Galphas. J Biol Chem 284(46):31714–31725. doi:10.1074/jbc.M109.028100
Chhangani D, Jana NR, Mishra A (2013) Misfolded proteins recognition strategies of E3 ubiquitin ligases and neurodegenerative diseases. Mol Neurobiol 47(1):302–312. doi:10.1007/s12035-012-8351-0
Matz A, Lee SJ, Schwedhelm-Domeyer N, Zanini D, Holubowska A, Kannan M, Farnworth M, Jahn O et al (2015) Regulation of neuronal survival and morphology by the E3 ubiquitin ligase RNF157. Cell Death Differ 22(4):626–642. doi:10.1038/cdd.2014.163
Kim JH, Kim WT (2013) The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol 162(3):1733–1749. doi:10.1104/pp.113.220103
Guerra DD, Pratelli R, Kraft E, Callis J, Pilot G (2013) Functional conservation between mammalian MGRN1 and plant LOG2 ubiquitin ligases. FEBS Lett 587(21):3400–3405. doi:10.1016/j.febslet.2013.08.045
Jiao J, Sun K, Walker WP, Bagher P, Cota CD, Gunn TM (2009) Abnormal regulation of TSG101 in mice with spongiform neurodegeneration. Biochim Biophys Acta 1792(10):1027–1035. doi:10.1016/j.bbadis.2009.08.009
Borden KL (2000) RING domains: master builders of molecular scaffolds? J Mol Biol 295(5):1103–1112. doi:10.1006/jmbi.1999.3429
Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A 96(20):11364–11369
Borden KL, Freemont PS (1996) The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol 6(3):395–401
Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS et al (2014) A draft map of the human proteome. Nature 509(7502):575–581. doi:10.1038/nature13302
Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M et al (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144(1):67–78. doi:10.1016/j.cell.2010.11.050
Dobson CM (2004) Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol 15(1):3–16. doi:10.1016/j.semcdb.2003.12.008
Muchowski PJ, Wacker JL (2005) Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6(1):11–22. doi:10.1038/nrn1587
Walker LC, Diamond MI, Duff KE, Hyman BT (2013) Mechanisms of protein seeding in neurodegenerative diseases. JAMA Neurol 70(3):304–310. doi:10.1001/jamaneurol.2013.1453
Demand J, Alberti S, Patterson C, Hohfeld J (2001) Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol 11(20):1569–1577
Chhangani D, Nukina N, Kurosawa M, Amanullah A, Joshi V, Upadhyay A, Mishra A (2014) Mahogunin ring finger 1 suppresses misfolded polyglutamine aggregation and cytotoxicity. Biochim Biophys Acta. doi:10.1016/j.bbadis.2014.04.014
Jacomy H, Talbot PJ (2003) Vacuolating encephalitis in mice infected by human coronavirus OC43. Virology 315(1):20–33
Fraser JR (2002) What is the basis of transmissible spongiform encephalopathy induced neurodegeneration and can it be repaired? Neuropathol Appl Neurobiol 28(1):1–11
Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L (2007) Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell 18(4):1129–1142. doi:10.1091/mbc.E06-09-0787
Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I et al (2004) Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev 18(14):1737–1752. doi:10.1101/gad.294904
Li L, Liao J, Ruland J, Mak TW, Cohen SN (2001) A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc Natl Acad Sci U S A 98(4):1619–1624. doi:10.1073/pnas.98.4.1619
Cheng TH, Cohen SN (2007) Human MDM2 isoforms translated differentially on constitutive versus p53-regulated transcripts have distinct functions in the p53/MDM2 and TSG101/MDM2 feedback control loops. Mol Cell Biol 27(1):111–119. doi:10.1128/MCB.00235-06
Sun K, Johnson BS, Gunn TM (2007) Mitochondrial dysfunction precedes neurodegeneration in mahogunin (Mgrn1) mutant mice. Neurobiol Aging 28(12):1840–1852. doi:10.1016/j.neurobiolaging.2007.07.012
Miller KA, Gunn TM, Carrasquillo MM, Lamoreux ML, Galbraith DB, Barsh GS (1997) Genetic studies of the mouse mutations mahogany and mahoganoid. Genetics 146(4):1407–1415
Prusiner SB (1991) Molecular biology of prion diseases. Science 252(5012):1515–1522
Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z et al (1993) Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A 90(23):10962–10966
Miesbauer M, Rambold AS, Winklhofer KF, Tatzelt J (2010) Targeting of the prion protein to the cytosol: mechanisms and consequences. Curr Issues Mol Biol 12(2):109–118
Chakrabarti O, Hegde RS (2009) Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell 137(6):1136–1147. doi:10.1016/j.cell.2009.03.042
Silvius D, Pitstick R, Ahn M, Meishery D, Oehler A, Barsh GS, DeArmond SJ, Carlson GA et al (2013) Levels of the Mahogunin Ring Finger 1 E3 ubiquitin ligase do not influence prion disease. PLoS One 8(1), e55575. doi:10.1371/journal.pone.0055575
Uchiyama K, Muramatsu N, Yano M, Usui T, Miyata H, Sakaguchi S (2013) Prions disturb post-Golgi trafficking of membrane proteins. Nat Commun 4:1846. doi:10.1038/ncomms2873
Yuan F, Yang L, Zhang Z, Wu W, Zhou X, Yin X, Zhao D (2013) Cellular prion protein (PrPC) of the neuron cell transformed to a PK-resistant protein under oxidative stress, comprising main mitochondrial damage in prion diseases. J Mol Neurosci : MN 51(1):219–224. doi:10.1007/s12031-013-0008-6
Winkler J, Tyedmers J, Bukau B, Mogk A (2012) Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 198(3):387–404. doi:10.1083/jcb.201201074
Lin Z, Zhao D, Yang L (2013) Interaction between misfolded PrP and the ubiquitin-proteasome system in prion-mediated neurodegeneration. Acta Biochim Biophys Sin (Shanghai) 45(6):477–484. doi:10.1093/abbs/gmt020
Liberski PP, Brown DR, Sikorska B, Caughey B, Brown P (2008) Cell death and autophagy in prion diseases (transmissible spongiform encephalopathies). Folia Neuropathol 46(1):1–25
Du Toit A (2014) Post-translational modification: sweetening protein quality control. Nat Rev Mol Cell Biol 15(5):295. doi:10.1038/nrm3787
van Oosten-Hawle P, Morimoto RI (2014) Transcellular chaperone signaling: an organismal strategy for integrated cell stress responses. J Exp Biol 217(Pt 1):129–136. doi:10.1242/jeb.091249
Scotter EL, Vance C, Nishimura AL, Lee YB, Chen HJ, Urwin H, Sardone V, Mitchell JC et al (2014) Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci 127(Pt 6):1263–1278. doi:10.1242/jcs.140087
Sontag EM, Vonk WI, Frydman J (2014) Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol 26:139–146. doi:10.1016/j.ceb.2013.12.006
Wolff S, Weissman JS, Dillin A (2014) Differential scales of protein quality control. Cell 157(1):52–64. doi:10.1016/j.cell.2014.03.007
Polling S, Mok YF, Ramdzan YM, Turner BJ, Yerbury JJ, Hill AF, Hatters DM (2014) Misfolded polyglutamine, polyalanine, and superoxide dismutase 1 aggregate via distinct pathways in the cell. J Biol Chem 289(10):6669–6680. doi:10.1074/jbc.M113.520189
Mosser DD, Caron AW, Bourget L, Denis-Larose C, Massie B (1997) Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol Cell Biol 17(9):5317–5327
Kakkar V, Meister-Broekema M, Minoia M, Carra S, Kampinga HH (2014) Barcoding heat shock proteins to human diseases: looking beyond the heat shock response. Dis Model Mech 7(4):421–434. doi:10.1242/dmm.014563
Chhangani D, Mishra A (2013) Mahogunin ring finger-1 (MGRN1) suppresses chaperone-associated misfolded protein aggregation and toxicity. Sci Rep 3:1972. doi:10.1038/srep01972
Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171(4):603–614. doi:10.1083/jcb.200507002
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282(33):24131–24145. doi:10.1074/jbc.M702824200
Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M et al (2002) p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol 160(1):255–263. doi:10.1016/S0002-9440(10)64369-6
Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M (2000) Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem 275(21):16023–16029
Ishii T, Yanagawa T, Yuki K, Kawane T, Yoshida H, Bannai S (1997) Low micromolar levels of hydrogen peroxide and proteasome inhibitors induce the 60-kDa A170 stress protein in murine peritoneal macrophages. Biochem Biophys Res Commun 232(1):33–37. doi:10.1006/bbrc.1997.6221
Viiri J, Hyttinen JM, Ryhanen T, Rilla K, Paimela T, Kuusisto E, Siitonen A, Urtti A et al (2010) p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol Vis 16:1399–1414
Robitaille Y, Lopes-Cendes I, Becher M, Rouleau G, Clark AW (1997) The neuropathology of CAG repeat diseases: review and update of genetic and molecular features. Brain Pathol 7(3):901–926
Donaldson KM, Li W, Ching KA, Batalov S, Tsai CC, Joazeiro CA (2003) Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc Natl Acad Sci U S A 100(15):8892–8897. doi:10.1073/pnas.1530212100
McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y et al (2000) CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet 9(14):2197–2202
Perez MK, Paulson HL, Pendse SJ, Saionz SJ, Bonini NM, Pittman RN (1998) Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol 143(6):1457–1470
Yamanaka T, Nukina N (2010) Transcription factor sequestration by polyglutamine proteins. Methods Mol Biol 648:215–229. doi:10.1007/978-1-60761-756-3_14
Sugars KL, Rubinsztein DC (2003) Transcriptional abnormalities in Huntington disease. Trends Genet 19(5):233–238. doi:10.1016/S0168-9525(03)00074-X
Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP et al (2000) The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A 97(12):6763–6768. doi:10.1073/pnas.100110097
Yamanaka T, Miyazaki H, Oyama F, Kurosawa M, Washizu C, Doi H, Nukina N (2008) Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J 27(6):827–839. doi:10.1038/emboj.2008.23
Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR et al (2005) CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J Neurosci : Off J Soc Neurosci 25(40):9152–9161. doi:10.1523/JNEUROSCI.3001-05.2005
Ying Z, Wang H, Fan H, Zhu X, Zhou J, Fei E, Wang G (2009) Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum Mol Genet 18(22):4268–4281. doi:10.1093/hmg/ddp380
Yang H, Zhong X, Ballar P, Luo S, Shen Y, Rubinsztein DC, Monteiro MJ, Fang S (2007) Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp Cell Res 313(3):538–550. doi:10.1016/j.yexcr.2006.10.031
Mishra A, Dikshit P, Purkayastha S, Sharma J, Nukina N, Jana NR (2008) E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J Biol Chem 283(12):7648–7656. doi:10.1074/jbc.M706620200
Barsh G, Gunn T, He L, Schlossman S, Duke-Cohan J (2000) Biochemical and genetic studies of pigment-type switching. Pigment Cell Res 13(Suppl 8):48–53
Hida T, Wakamatsu K, Sviderskaya EV, Donkin AJ, Montoliu L, Lynn Lamoreux M, Yu B, Millhauser GL et al (2009) Agouti protein, mahogunin, and attractin in pheomelanogenesis and melanoblast-like alteration of melanocytes: a cAMP-independent pathway. Pigment Cell Melanoma Res 22(5):623–634. doi:10.1111/j.1755-148X.2009.00582.x
Barsh GS (2007) Regulation of pigment type switching by agouti, melanocortin signaling, attractin, and mahoganoid. In: The Pigmentary System: Physiology and Pathophysiology: Second Edition. pp 395–409. doi:10.1002/9780470987100.ch19
Walker WP, Gunn TM (2009) Piecing together the pigment-type switching puzzle. Pigment Cell Melanoma Res 23(1):4–6. doi:10.1111/j.1755-148X.2009.00654.x
Bahr BA, Bendiske J (2002) The neuropathogenic contributions of lysosomal dysfunction. J Neurochem 83(3):481–489
Kurz T, Terman A, Gustafsson B, Brunk UT (2008) Lysosomes in iron metabolism, ageing and apoptosis. Histochem Cell Biol 129(4):389–406. doi:10.1007/s00418-008-0394-y
Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA (2008) Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci : Off J Soc Neurosci 28(27):6926–6937. doi:10.1523/JNEUROSCI.0800-08.2008
Kirkegaard T, Roth AG, Petersen NH, Mahalka AK, Olsen OD, Moilanen I, Zylicz A, Knudsen J et al (2010) Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature 463(7280):549–553. doi:10.1038/nature08710
Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C (2014) The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab 19(3):357–372. doi:10.1016/j.cmet.2014.01.010
Beal MF (2005) Mitochondria take center stage in aging and neurodegeneration. Ann Neurol 58(4):495–505. doi:10.1002/ana.20624
Collignon J, Varlet I, Robertson EJ (1996) Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381(6578):155–158. doi:10.1038/381155a0
Lowe LA, Supp DM, Sampath K, Yokoyama T, Wright CV, Potter SS, Overbeek P, Kuehn MR (1996) Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature 381(6578):158–161. doi:10.1038/381158a0
Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H (1996) Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature 381(6578):151–155. doi:10.1038/381151a0
Shiratori H, Hamada H (2006) The left-right axis in the mouse: from origin to morphology. Development 133(11):2095–2104. doi:10.1242/dev.02384
Yan YT, Gritsman K, Ding J, Burdine RD, Corrales JD, Price SM, Talbot WS, Schier AF et al (1999) Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev 13(19):2527–2537
Cota CD, Bagher P, Pelc P, Smith CO, Bodner CR, Gunn TM (2006) Mice with mutations in Mahogunin ring finger-1 (Mgrn1) exhibit abnormal patterning of the left-right axis. Dev Dyn 235(12):3438–3447. doi:10.1002/dvdy.20992
Menzies FM, Fleming A, Rubinsztein DC (2015) Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci 16(6):345–357. doi:10.1038/nrn3961
Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292(5521):1552–1555. doi:10.1126/science.292.5521.1552
Nau R, Sorgel F, Eiffert H (2010) Penetration of drugs through the blood-cerebrospinal fluid/blood–brain barrier for treatment of central nervous system infections. Clin Microbiol Rev 23(4):858–883. doi:10.1128/CMR.00007-10
Pardridge WM (2011) Drug transport in brain via the cerebrospinal fluid. Fluids and barriers of the CNS 8(1):7. doi:10.1186/2045-8118-8-7
Ohtsuka K, Kawashima D, Gu Y, Saito K (2005) Inducers and co-inducers of molecular chaperones. Int J Hyperth 21(8):703–711. doi:10.1080/02656730500384248
Nagai Y, Fujikake N, Popiel HA, Wada K (2010) Induction of molecular chaperones as a therapeutic strategy for the polyglutamine diseases. Curr Pharm Biotechnol 11(2):188–197
Rokutan K (2003) Molecular chaperone inducers in medicine and diseases. Nihon Yakurigaku Zasshi 121(1):15–20
Lilienbaum A (2013) Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol 4(1):1–26
Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM (2007) Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 171(2):513–524. doi:10.2353/ajpath.2007.070188
Chhangani D, Chinchwadkar S, Mishra A (2014) Autophagy coupling interplay: can improve cellular repair and aging? Mol Neurobiol 49(3):1270–1281. doi:10.1007/s12035-013-8599-z
Raina K, Crews CM (2010) Chemical inducers of targeted protein degradation. J Biol Chem 285(15):11057–11060. doi:10.1074/jbc.R109.078105
Stone S, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137(1):13–30. doi:10.1104/pp.104.052423
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi:10.1038/msb.2011.75
Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20(3):426–427. doi:10.1093/bioinformatics/btg430
Cooray SN, Guasti L, Clark AJ (2011) The E3 ubiquitin ligase Mahogunin ubiquitinates the melanocortin 2 receptor. Endocrinology 152(11):4224–4231. doi:10.1210/en.2011-0147
Gunn TM, Silvius D, Bagher P, Sun K, Walker KK (2013) MGRN1-dependent pigment-type switching requires its ubiquitination activity but not its interaction with TSG101 or NEDD4. Pigment Cell Melanoma Res 26(2):263–268. doi:10.1111/pcmr.12059
Chhangani D, Nukina N, Kurosawa M, Amanullah A, Joshi V, Upadhyay A, Mishra A (2014) Mahogunin ring finger 1 suppresses misfolded polyglutamine aggregation and cytotoxicity. Biochim Biophys Acta 1842(9):1472–1484. doi:10.1016/j.bbadis.2014.04.014
Srivastava D, Chakrabarti O (2014) Mahogunin-mediated alpha-tubulin ubiquitination via noncanonical K6 linkage regulates microtubule stability and mitotic spindle orientation. Cell Death Dis 5, e1064. doi:10.1038/cddis.2014.1
Cheng D, Xiong C, Li J, Sui C, Wang S, Li H, Jiang X (2014) The effect of mahogunin gene mutant on reproduction in male mice: a new sight for infertility? Andrologia 46(2):98–105. doi:10.1111/and.12050
Acknowledgments
This work was supported by the Department of Biotechnology, Government of India. AM was supported by Ramalinganswami Fellowship (BT/RLF/Reentry/11/2010) and Innovative Young Biotechnologist Award (IYBA) scheme (BT/06/IYBA/2012) from the Department of Biotechnology, Government of India. AU was supported by a research fellowship from the Council of Scientific and Industrial Research-University Grants Commission (CSIR-UGC), Government of India. The authors would like to thank Mr. Bharat Pareek for his technical assistance and the entire lab management during the manuscript preparation. We apologize to various authors whose work could not be included due to space limitations.
Compliance with Ethical Requirements
ᅟ
Conflict of Interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Upadhyay, A., Amanullah, A., Chhangani, D. et al. Mahogunin Ring Finger-1 (MGRN1), a Multifaceted Ubiquitin Ligase: Recent Unraveling of Neurobiological Mechanisms. Mol Neurobiol 53, 4484–4496 (2016). https://doi.org/10.1007/s12035-015-9379-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-015-9379-8