Abstract
Aims
Plants host communities of fungal and bacterial endophytes, establishing a complex network of multipartite interactions, but the mechanisms whereby they interact are poorly understood. Some fungi, such as the beneficial mycorrhiza-like fungus Serendipita (=Piriformospora) indica, can be helped by bacteria for establishment, survival and colonization. Although this fungus harbors a Rhizobium as an endofungal bacterium, we hypothesized that other bacteria might also establish associations with the fungus and combining S. indica with bacteria might enhance plant growth and health.
Methods
The interactions among S. indica and four endophytic Proteobacteria belonging to Methylobacterium, Tardiphaga, Rhodanobacter and Trinickia spp. were characterized in vitro and for their effect on tomato growth and biocontrol of Fusarium oxysporum and Rhizoctonia solani. Possible mechanisms behind these interactions were described based on genome and microscopic analyses, using fungal and bacterial strains tagged with fluorescent markers.
Results
All bacteria stimulated S. indica growth in vitro. Moreover, several of the bacteria stimulated growth of tomato plants, but co-inoculations with S. indica and bacteria did not perform better than single inoculations. Contrarily, combinations of S. indica and bacteria significantly reduced disease progression of fungal pathogens. These microbes seem to cooperate in the process of root colonization for instance by increasing fungal sporulation and hyphae expansion, showing multipartite interaction between microbes and plants. Interestingly, the strain of Trinickia internally colonizes spores of S. indica as an endofungal bacterium during in vitro-co-culturing, suggesting further that the fungus might acquire formerly unrecognized genera of bacteria and genome analysis of the bacteria revealed many genes potentially involved in fungal and plant growth stimulation, biocontrol and root colonization, highlighting putative mechanisms of plant-fungal-bacterial interaction.
Conclusions
Our study represents an important step towards unraveling the complex interactions among plants, S. indica, endophytic bacteria and fungal pathogens, and indicates that adding bacteria to fungal inoculum could have a remarkable impact on the plant-S. indica symbiosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Arbuscular mycorrhizal fungi (AMF) and rhizobacteria are among the most abundant components of the plant microbiota, and hence crucial drivers of plant evolution. They thrive in the plant rhizosphere in all natural and managed ecosystems but can also penetrate the roots and establish as endophytes (Compant et al. 2005b). In both niches, these microorganisms can improve plant nutrition, increase tolerance to abiotic stresses and enhance protection against phytopathogens (Bonfante and Anca 2009; Martínez-Arias et al. 2019). Some bacteria are capable of boosting colonization and establishment of endophytic and mycorrhizal fungi, and thus they have been regarded as mycorrhizal helper bacteria (Garbaye 1994; Frey-Klett et al. 2007). The physical connection between them can range from apparently disordered microbial assemblies to specific symbiotic associations of fungal hyphae and bacterial cells (Frey-Klett et al. 2011). Already in 1970, endofungal bacteria were detected living inside hyphae and spores of the AM fungus Endogone (Mosse 1970). Since then, several genera of bacteria have been identified residing inside mycorrhizal fungi (Bianciotto et al. 1996; Hoffman and Arnold 2010) and seem to play a crucial role in the mycorrhizal symbiosis with plants (Partida-Martinez and Hertweck 2005; Salvioli et al. 2016). However, the complexity of the fungal-bacterial-plant interactions is still far from being fully elucidated. Understanding the functioning of the plant microbiome is an essential step for the exploitation of beneficial microorganisms, both fungi and bacteria, for the improvement of crop productivity and biological control of plant pathogens.
Serendipita indica, formerly known as Piriformospora indica, is an AM-like fungus originally isolated in the Indian Thar desert (Verma et al. 1998). It can endophytically colonize a wide range of plants, but unlike mycorrhizal fungi, this fungus depends on the host cell death for successful colonization (Deshmukh et al. 2006). S. indica promotes plant growth, induces tolerance to abiotic stresses, and confer resistance to shoot and root pathogens (Waller et al. 2005) by hormone-manipulation of the host plant, enhancement of induced systemic resistance (ISR) and activation of the antioxidative system (reviewed in Franken 2012; and Gill et al. 2016). Intriguingly, this fungus hosts an endofungal bacterium, Rhizobium radiobacter F4 (Sharma et al. 2008). In comparison to obligate endofungal bacteria, the genome of RrF4 is not reduced and the bacterium is able to live independently of its host and to thrive as an endophyte, thus forming a facultative symbiosis with the fungus (Glaeser et al. 2016). It hints at the interaction is not specific and other bacteria might also colonize and establish a facultative symbiosis with S. indica. Moreover, the endofungal bacterium RrF4 possesses plant growth-promoting properties and the ability to confer resistance to fungal plant pathogens. Therefore, it has been hypothesized that some of the beneficial properties attributed to S. indica might stem from the effect exerted by the bacterium (Qiang et al. 2012a; Glaeser et al. 2016).
It appears likely that other bacteria with plant growth-promoting (PGP) traits might also contribute to further increase fungal fitness and the PGP and biocontrol effects reported by this fungus. For instance, the model bacterium Bacillus subtilis and the strain WR5 of Azotobacter chrococcum were shown to enhance the growth and sporulation of S. indica in vitro (Jiang et al. 2018; Bhuyan et al. 2015). Some researchers have also investigated the effect of combining S. indica with plant growth-promoting rhizobacteria (PGPR) on plant growth: co-inoculation of S. indica with the phosphate-solubilizing bacterium Pseudomonas striata increased plant growth in chickpea (Meena et al. 2010), and a talcum-based formulation of S. indica and Pseudomonas sp. R81 enhanced plant biomass in tomato (Sarma et al. 2011). However, it has also been reported that dual inoculations of S. indica with Paenibacillus or pseudomonads did not improve (Nautiyal et al. 2010) or even displayed lower plant dry weight than single inoculations (Sarma et al. 2011). One way of improving biocontrol may be to combine microbes, especially if they exhibit complementary modes of action and abilities to colonize root microsites (Whipps 2001). Although some combinations of bacteria and fungi have demonstrated to enhance biocontrol (Guetsky et al. 2001; Sarma et al. 2011), there are several studies in which combinations of various bacteria and fungi provided poorer biocontrol activities than the microbes used singly (Hervás et al. 1998; Xu et al. 2011). In light of these incompatibilities, there is a need to further investigate new fungal-bacterial synergisms and the mechanisms involved in this interplay for an efficient exploitation and engineering of the plant microbiome.
In a previous study, we detected several endophytic bacterial strains from very diverse families with the ability to stimulate S. indica growth in vitro. In that study, we focused on Mycolicibacterium strains, as this genus showed to comprise the highest number of strains having a beneficial effect on S. indica (del Barrio-Duque et al. 2019). However, strains from other taxa might have similar beneficial properties on S. indica but potentially employing different mechanisms. Among those bacteria showing the most outstanding stimulatory effects on S. indica growth, we selected four proteobacterial strains of Methylobacterium sp., Tardiphaga sp., Rhodanobacter sp., and Trinickia (=Paraburkholderia) sp. The uniquely reported endofungal bacterium of S. indica (RrF4) belongs to the proteobacterial order Rhizobiales (Sharma et al. 2008), and perhaps other bacteria belonging to Rhizobiales, like Methylobacterium or Tardiphaga, might also establish a synergistic interaction with S. indica. Members of the order Burkholderiales (e.g. Trinickia) are among the most frequently identified endofungal bacteria (Compant et al. 2008). Bacteria of the genus Rhodanobacter (Xanthomonadales) seem further to be implicated in shaping communities of AMF (Svenningsen et al. 2018) and several members of the same order have been considered as ‘fungiphile’ bacteria (Simon et al. 2015). These features make the selected strains potential candidates for studying fungal-bacterial interactions. Moreover, the four selected strains are phylogenetically closely related to other strains of PGPR described to enhance plant growth or to be involved in the biocontrol of pathogens (Tani et al. 2015; Berg 2009; De Clercq et al. 2006; Sessitsch et al. 2005). We therefore hypothesized that strains belonging to these taxa may also influence the fitness, establishment and colonization of S. indica and enhance the effects of this symbiotic fungus on plant growth and biocontrol. We further speculated that they might internally colonize S. indica and behave as endofungal bacteria.
Materials and methods
Microorganisms
Serendipita (=Piriformospora) indica strain DSM 11827 (Varma et al. 1999) was provided by Prof. Philipp Franken (Erfurt Research Centre for Horticultural Crops, Germany). The fungus was kept at −80 °C in a mixture 3:1 of Potato Dextrose Broth (PDB) (Carl Roth, Germany) and glycerol, and grown on Potato Dextrose Agar (PDA) plates or in liquid culture containing Aspergillus complete medium (Pontecorvo et al. 1953).
Rhizoctonia solani AG-3 was provided by Dr. Rosanna C. Hennessy (University of Copenhagen, Denmark) and Fusarium oxysporum f.sp. lycopersici Fol4287 by Maria E. Constantin (University of Amsterdam, Netherlands) (Di Pietro and Roncero 1996). Both fungal pathogens were maintained at −80 °C in a mixture of 3:1 PDB + glycerol.
The endophytic bacterial strains Methylobacterium sp. P1–11, Tardiphaga sp. P9–11, Rhodanobacter sp. T12–5, and Trinickia sp. T12–10 were obtained from the AIT (Austrian Institute of Technology, Tulln, Austria) strain collection. They were isolated from tomato (T) or potato (P) roots (del Barrio-Duque et al. 2019).
Effect of bacterial strains on mycelial growth of S. indica and fungal pathogens
The growth of the beneficial fungus S. indica or the pathogens F. oxysporum and R. solani in interaction with bacteria was assessed by measuring hyphae development on agar plates as described in del Barrio-Duque et al. (2019). For these experiments, bacteria were precultured for four days on Nutrient Agar No2 (NA) (Sigma-Aldrich, St. Louis, USA) and then streaked individually on 1 cm2 in the center of a Petri dish (9 cm Ø) containing 15 ml of PDA or a mixture 1:1 of PDA + NA. S. indica was precultured for two weeks on PDA and then a 0.5 cm2 agar plug of active mycelium was placed upside down onto the streaked bacterium. Additionally, S. indica was co-cultured on PDA with bacteria streaked 4 cm away from the agar plug. All the co-cultures were replicated four times. As a control, the fungus was grown alone. Plates were incubated in darkness at 26 °C and mycelial growth was monitored after 3, 6, 9 and 11 days of dual-culturing, depending on the growth rate of each fungus. The surface of the Petri dish covered with mycelium was then quantified with ImageJ 1.48 software (https://imagej.nih.gov/ij/).
For estimation of dry fungal biomass and spore yield during interactions with bacteria, seven PDA plugs from the periphery of a two-week old culture of S. indica were transferred to 100 ml Erlenmeyer flasks with 25 ml PDB and incubated at 26 °C and 150 rpm. After six days, 1 ml of a bacterial culture grown on Nutrient Broth (NB) (Difco, Detroit, MI) for two days (OD 0.3 at 630 nm) was added to each flask and co-cultured for additional four days. Mycelium was then washed thrice with distilled water and collected by centrifugation (Thermo Scientific Heraeus Megafuge 40R) at 4600 rpm for 5 min. Dry fungal weight was determined after drying the mycelium in the oven at 60 °C for three days. Spore production was estimated in a hemocytometer after grinding and sonicating the fungal pellet according to Kumar et al. (2011).
Co-inoculation of S. indica and endophytic bacteria for tomato growth promotion
To produce fungal inoculum, agar plugs from the active mycelium of a 2-week old PDA culture of S. indica were transferred to 250 ml Erlenmeyer flasks containing 100 ml of Aspergillus complete medium and incubated at 26 °C for 3 weeks at 150 rpm in a shaking incubator. Mycelium and spores were harvested by centrifugation (4500 rpm, 5 min) and washed 3 to 5 times with sterile phosphate-buffered saline (PBS) of pH 6.5. The mixture of mycelium and spores was crushed with a homogenizer Ultra Turrax T25 (IKA®, Staufen, Germany) in intervals of 30 s for 3 min. Spores + mycelium fragments were measured with a hemocytometer (NanoEnTek, Seoul, Korea) and the viability of the spores + mycelium fragments was confirmed by plating on PDA. Final inoculum ratios were adjusted with PBS.
For bacteria, they were grown for 60 h in round-bottom Falcon tubes containing 5 ml of NB (26 °C, 190 rpm), then centrifuged (4600 rpm, 6 min, room T°C) and washed thrice with sterile PBS to discard traces of media. Bacterial growth and CFU were preliminarily assessed by measuring the optical density at 630 nm and plating standard serial dilutions on NA, then the amount of inoculum was calibrated to the final concentration of bacterial cells with PBS.
For plant experiments, tomato seeds cv. “Moneymaker” (Austrosaat, Vienna, Austria) were germinated in a Whatman® filter paper (110 mm Ø) for 4 days and then transferred to Falcon tubes (50 ml) containing 15 ml of PBS and either i) spores + hyphae fragments of S. indica (5 × 105 CFU/ml), ii) bacteria (5 × 107 CFU/ml or iii) a blend of S. indica (5 × 105 CFU/ml) with bacteria (5 × 107 CFU/ml). Falcon tubes were kept for 30 min in a horizontal Tube Roller RS-TR5 (Phoenix Instrument GmbH, Garbsen, Germany). Control seeds were immersed in 1xPBS. Seeds were then sown in 1-l pots containing the substrate “Fruhstorfer Erde Typ Nullerde” (Hawita Gruppe, Vechta, Germany) at 1 cm depth. The plants were grown in the greenhouse with a 12 h light/dark photoperiod, day/night temperature of 22/21 °C, and a relative humidity 50/35%, and watered twice a week with tap water. Plants were harvested 6 weeks after planting and shoot fresh weight and leaf area were measured (using ImageJ software). The experiment consisted of 10 treatments; ± S. indica and ± bacteria (four strains) with 14 replicates (7 pots × 2 plants per pot) for each treatment. Additionally, the experiment was repeated with a high-nutrient soil composed of a mixture (1:1:1 v/v) of substrate “Tonsubstrat ED63 Special” (Einheitserde, Germany), perlite and sand. After harvesting, shoot fresh and dry weight (after oven-drying for 3 days at 70 °C) were measured.
Biocontrol of F. oxysporum and R. solani
Protection against F. oxysporum was determined by pot experiments in which tomato plants (cv. Moneymaker) were infected with the pathogen and with single or combined inoculations of S. indica and bacteria. Fusarium inoculum was produced according to Constantin et al. (2019). For this, a PDA plug from a 6-day old culture of Fusarium was transferred to 100 ml minimal media (3% sucrose, 1% KNO3 and 0.17% Yeast Nitrogen Base without ammonia and amino acids) and incubated for 5 days in the dark (26 °C, 190 rpm). Spores were separated with a Miracloth filter (Millipore), rinsed twice with sterile PBS and adjusted to 107 spores/ml. Bacterial and S. indica inoculum production as well as seed inoculation were performed as explained above. Inoculated seeds were then planted in 1 l pots containing a mixture (1:1:1 v/v) of the substrate “Tonsubstrat ED63 Special” (Einheitserde, Germany), perlite and sand. Ten replicated plants were assessed per treatment (5 pots × 2 seeds per pot). Ten-day-old tomato seedlings were uprooted, trimmed (leaving around 1 cm of root) to assist Fusarium infection (de Lamo et al. 2018) and placed in the Fusarium spore suspension for 30 min, and then immediately repotted. After five weeks, shoot fresh weight and the extent of disease progression were measured. Disease index (DI) was scored as described before (Gawehns et al. 2014) but adding DI = 5 when plants were dead. Briefly, DI 0 = no symptoms; 1 = one brown vessel above the soil; 2 = one-two brown vascular bundles at the cotyledon level; 3 = three or more brown vessels and growth deformation, 4 = all vessels brown or the plant is dwarfed and wilted, 5 = dead plant.
The effects of combinations of individual bacteria and S. indica against R. solani were determined by a germination test in sterile closed systems. Transparent Steri Vent Containers (107 × 94 × 96 mm, Duchefa Biochemie b.v, Haarlem, Netherlands) were filled with 120 g of a sterile (2x, 20 min, 121 °C) mixture (4:1, w/w) of distilled water and vermiculite (2–3 mm, Sigma-Aldrich). Tomato seeds were surface-sterilized for 5 min with 2.5% sodium hypochlorite and washed eight times with sterile water. Seeds were inoculated with ± S. indica and ± individual bacterial strains as earlier described. Five 5 mm plugs of a 3-week-old culture of Rhizoctonia grown in 1/5 PDA were placed in a row in the center of each container, and 2 rows of tomato seeds (5 seeds per row) were sown in both sides of the pathogen row 2 cm away from the pathogen. Control was prepared with 1/5 PDA plugs. The seeds and the Rhizoctonia agar plugs remained at 5 mm depth. Three replicated containers were included per treatment and kept in the greenhouse (see above) for 4 months. Germination and disease progression of the seedlings were monitored regularly and assessed by counting seedlings as follows: 1 = Plant germinated and no Rhizoctonia symptoms; 0.5 = Plant germinated and alive, but with necrotic areas in stem and leaves; 0 = dead plant.
Bacterial genome sequencing, assembly and annotation
To determine genome contents of the four Proteobacteria strains, bacterial genomic DNA was extracted with a phenol-chloroform based protocol. Briefly, bacteria were cultivated for 3 days in NB and harvested by centrifugation. The bacterial pellets were then resuspended in lysis buffer (0.2 mg/mL Proteinase K, 50 mM Tris-Cl, 1% SDS, 5 mM EDTA pH 8, 0.5 M NaCl,) and incubated overnight (65 °C, 400 rpm). DNA was extracted 2 times using 1 volume of phenol-chloroform-isoamylalcohol (25:24:1) and collected by centrifugation (5000 rpm, 2 min). Genomic DNA was then purified with Amicon Ultra 0.5 mL 30 K Centrifugal Filter Units (Millipore, Cork, Ireland) and resuspended in water. Whole-genome shotgun sequencing was performed on an Illumina HiSeq (GATC Biotech, Konstanz, Germany), producing 2 × 150 bp reads.
Illumina reads were screened for the presence of PhiX using Bowtie 2 (v2.3.4.3) (Langmead and Salzberg 2012) and adapters were trimmed with fastp (v0.19.5) (Chen et al. 2018b). Sequence length distribution and quality were checked via FastQC (Andrews 2010). Genome assembly was performed with SPAdes v3.13.0 (Bankevich et al. 2012) and low-abundant (< 2x), short (< 500 bp) contigs discarded. Contaminant contigs were examined using BlobTools and alien contigs were eventually filtered out. Genome assembly quality was inferred using QualiMap v2.2 (Okonechnikov et al. 2015) and QUAST v5.0.0 (Gurevich et al. 2013) and then genome completeness reconstruction was evaluated with CheckM v1.0.18 (Parks et al. 2015). Gene annotation was carried out using Prokka v1.12 (Seemann 2014) and NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016). The presence of plasmids was ascertained by using Mash v2.1 against the PLSDB database (Galata et al. 2018). Putative plasmid contigs were screened for the presence of genes for replication initiator proteins, repA. For this purpose, a curated FASTA file with ~8000 repA genes was generated from plasmidial genome sequences in NCBI. These repA gene sequences were used to build a database against which the selected contigs were BLASTed. Functional annotation was performed using the ClassicRAST (Rapid Annotation using Subsystem Technology) web server (http://rast.nmpdr.org) (Aziz et al. 2008) and the hierarchical orthology framework EggNOG 4.5 (Huerta-Cepas et al. 2015). CAZy families were ascertained with dbCAN2 based on the DIAMOND database. A cutoff of E-Value of 1e-102 was set for the output. In the event of a gene ambiguously determined as CBM plus other CAZy classes, the gene was classified as CBM. Annotation of proteins was based on the CAZy database (Lombard et al. 2013). Biosynthetic gene clusters and secondary metabolites were further predicted using antiSMASH version 4.0.2 (Weber et al. 2015). Circular plots of genomes were generated, and decorated with important features, using BLAST Ring Image Generator (BRIG, version 0.95) (Alikhan et al. 2011). Default parameters were used for all software unless otherwise specified. Based on the functionality of certain genes previously reported in the literature and the functional annotation ascertained with these genomic analysis tools, genes were proposed as potential candidates to explain the observed phenotypic effects in planta and in vitro.
In order to assign objective taxonomic classifications to these genomes, the software toolkit GTDB-Tk v0.3.2 was employed (Chaumeil et al. 2019; Parks et al. 2018). Genomes were considered classified at species level when the Average Nucleotide Identity (ANI) value was within the circumscription radius of 95% and the sequence alignment fraction (AF) was > = 0.65. The draft genome sequences of the strains P1–11, P9–11, T12–5 and T12–10 are available at NCBI, BioProject PRJNA393298, with the DDBJ/ENA/GenBank accession numbers NPKS00000000, NPKQ00000000, NPKN00000000 and NPKM0000000 respectively.
Quantification of root colonization by S. indica
To study the effect of bacteria on the colonization of the roots by S. indica, tomato seedlings were grown in Transparent Steri Vent Containers as described above, but without addition of Rhizoctonia agar plugs, and inoculated with S. indica alone and in combination with bacterial strains P1–11 or T12–10 wild-types as earlier described. For every treatment, 3 seedlings from each transparent (×3) container were sampled 10 days and 8 weeks after planting and maintained at −20 °C. Roots were stained according to a modified protocol of Venneman et al. (2017). Briefly, roots were abundantly rinsed under running tap water and incubated at 80 °C in 10% KOH solution for 30 min, and then at room temperature for an additional 30 min. After a rinsing step, roots were acidified for 20 min at room temperature in 1% acetate solution before adding droplets of china blue ink (2 droplets per 25 ml). This mixture was incubated for 30 min at 80 °C and at room temperature for an additional 30 min. The roots were then rinsed with water and immersed in a glycerol solution for 10 min (140 ml glycerol +46 ml water +14 ml 1% acetate) before to be rinsed again. From every stained root, fragments of 1 cm length were cut and observed with a microscope Nikon Eclipse E200, and the presence of hyphae and spores of S. indica were quantified. In total 5 to 7 plantlets per treatment were analyzed to have 20 root sections to quantify.
Fluorescent labeling of the endophytic bacteria
The four strains were tagged with the red fluorescent marker mCherry by transformation with the self-replicating broad-host-range plasmid pSEVA237R (Silva-Rocha et al. 2013). This plasmid does not contain transposons and is not inserted chromosomally but can replicate itself in the recipient cell. Wild-type strains were grown in 10 ml of a PDB + NB mixture (1:1) in 50 ml Falcon tubes for 24 h in a shaking incubator (28 °C, 180 rpm). Cells were harvested by centrifugation (4700 rpm, 10 min, 4 °C), washed thrice with 300 mM sucrose, and then resuspended in 400 μl of 300 mM sucrose. One hundred μl of the competent cells were mixed with 500 ng of purified pSEVA237R plasmid. The mixture was electroporated with a Gene Pulser II (Bio-Rad, Richmond, Calif.) by a pulse of 25 μF, 200 Ω, 2.5 kV, and then 1 ml of PDB + NB was immediately added to the mixture. The mixture was incubated for 2 h at 28 °C in a shaking incubator and then plated on PDA + NA amended with 50 μg/ml kanamycin (Km). Colonies and cells of the mCherry-marked strains were examined by using a fluorescence microscope (Olympus model SZX16; Tokyo, Japan). Transformant stability was assessed by culturing tagged strains in PDB + NB with or without kanamycin for over five generations, and then plating cells on PDA + NA w/o antibiotics. Moreover, the growth pattern as well as colony and cell morphologies of wild-type and mCherry-tagged bacteria were compared.
Confocal laser scanning microscopy (CLSM) analysis
CLSM assays were performed with GFP-labelled S. indica (gift from Prof. Alga Zuccaro, University of Cologne, Germany) and the red fluorescent-labelled bacterial strains Methylobacterium sp. P1–11:mCherry and Trinickia sp. T12–10:mCherry that were successfully transformed. Interactions between the fungus and each bacterium were examined in Petri dishes and on roots. For the in vitro analysis, S. indica and bacterial strains were dual-cultured in direct contact on PDA as earlier described. No antibiotics were included for the bacterial preculturing. After 11 days of co-culturing, fragments of fungal mycelium were sampled with a scalpel from the surface of the agar plate, roughly 0.5 cm away from the spot at which the fungal agar plug and the bacteria were initially confronted. For each bacterial-fungal combination, samples were retrieved and observed from four replicated Petri dishes.
Aiming at investigating root colonization patterns of S. indica and bacteria, tomato seedlings were grown in Transparent Steri Vent Containers as described above. Inoculum production of transformed microbes was obtained with the same protocol as for the wild type strains, but to avoid loss of the plasmid-born fluorescence, the strain Methylobacterium sp. P1–11:mCherry (but not Trinickia sp. T12–10:mCherry) was grown on NB + 50 μg/ml Km. The seedlings were harvested 14 dap. For every treatment (± S. indica and ± individual bacterial strains) at least 3 seedlings from each transparent (×3) container were sampled and observed.
The samples were then examined under a confocal microscope (Olympus Fluoview FV1000 with multiline laser FV5-LAMAR-2 HeNe(G) and laser FV10-LAHEG230–2). X, Y, Z pictures were acquired with 20X, 40X or 60X objectives at 405, 488, 594 nm wavelengths and then merged (RGB) using the software of the microscope provider (Olympus FV1000). Confocal stacks are presented as single optical slices or maximum projections (Cardinale and Berg 2015). The light/contrast balance was improved to better observe the image details when image acquisition remained very dark (as described in Glassner et al. 2015). Three-dimensional reconstructions (Cardinale and Berg 2015) were modeled using the software Imaris® 7.2 (Bitplane AG, Zürich, Switzerland).
Statistical analysis
The data on in vitro bacterial- fungal dual-inoculations as well as plant biomass and leaf area from the growth enhancement experiments were analyzed in R 3.5.1 (Team RC 2019). Data distributions were evaluated with the fitdistrplus package (Delignette-Muller and Dutang 2015) and linear or linear mixed-effects models (nlme R package) (Pinheiro et al. 2019), when necessary, were created. After graphical validation of homogeneity assumption, ANOVA test was carried out on the generated models, followed by pairwise comparisons (Tukey’s method, P = 0.05) calculated by Estimated Marginal Means (emmeans R package) (Lenth 2019). Quantitative data were processed with dplyr package (Wickham et al. 2019) and displayed with boxplots using ggplot2 package (Wickham and Chang 2016). The data from the biocontrol experiments were analyzed using PRISM 8.0 (GraphPad, https://www.graphpad.com). Datasets from the germination test with Rhizoctonia were analyzed using one-way ANOVA and Tukey’s test (P = 0.05). Regarding the Fusarium assay, a non-parametric Mann–Whitney U-test was applied on the shoot fresh weight and disease index data (de Lamo et al. 2018; Constantin et al. 2019).
Results
Effect of bacterial strains on mycelial growth of S. indica and fungal pathogens
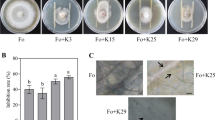
The four tested strains, Methylobacterium sp. P1–11, Tardiphaga sp. P9–11, Rhodanobacter sp. T12–5 and Trinickia sp. T12–10, significantly stimulated S. indica growth when co-cultured on PDA for all the measurements of hyphae expansion over the Petri dish (Fig. 1a-c and Suppl. Fig. 1). The stimulating effect was also observed on PDA + NA (Suppl. Fig. 1). When S. indica and the bacterial strain were placed at different spots of the Petri dish, the fungal growth was not impaired; rather the strains P1–11, P9–11 and T12–5 significantly promoted hyphal extension (Suppl. Fig. 1). In the experiments on liquid medium, strains P1–11 and T12–10 significantly increased fungal dry weight and sporulation (Fig. 1d-f). Strains P9–11 and T12–5 only exhibited a moderate non-significant increase in fungal biomass and spore yield, probably due to their slow growth in PDB.
Effect of bacterial strains on S. indica growth and sporulation. Growth of S. indica cultured alone (control) or in combination with bacterial strains after 3 (a) and 11 days (b-c) on PDA measured as mycelium expansion over the Petri dish (n = 4). Liquid cultures (PDB) of S. indica with bacterial strains after 10 days (d-f). Dry weight (d) and spore yield (e) of S. indica cultured alone (control) and in combination with bacteria (n = 3) on PDB. The boxplots show all data points, the horizontal line represents the median, surrounded by the upper (25th) and lower (75th) percentiles. Same letters represent non-significantly different mean values, according to Tukey’s test (P < 0.05), after ANOVA. Bar in the pictures corresponds to 4 cm
In contrast to the observed growth stimulation of S. indica, mycelial growth of the phytopathogen F. oxysporum and R. solani was not stimulated by bacteria (Suppl. Fig. 2). In contrast, the strains P1–11 on PDA, and T12–5 on PDA and PDA + NA, significantly reduced Fusarium growth, while the strain P9–11 significantly impaired the growth of Rhizoctonia on PDA + NA.
Co-inoculation of S. indica and endophytic bacteria for tomato growth promotion
Tomato plants received single or dual inoculations of S. indica and the bacterial strains. In no case microbial application impaired plant growth (Fig. 2a-c). Strain P1–11 only exhibited neutral effects and strain P9–11 yielded non-significant 2.0- and 4.2-fold increments of shoot fresh weight and leaf area respectively (Fig. 2b and c). Strains T12–5 and T12–10 significantly promoted plant growth, exhibiting 3.9- and 4.0-fold increments in shoot fresh weight, respectively (Fig. 2b). Inoculation with S. indica resulted also in notable increased fresh weight and leaf area. Dual inoculations of S. indica and bacteria significantly enhanced plant growth in comparison to control plants, however these combinations did not perform better than single inoculations. Interestingly, dual inoculation of S. indica + T12–5 displayed significant lower performance than inoculation of either microbe alone. This greenhouse trial was repeated in soil with a higher content of nutrients, but under these conditions no significant differences were detected between treatments of control plants and plants inoculated with microbes (Suppl. Fig. 3).
Effect of bacterial strains Methylobacterium sp. P1–11, Tardiphaga sp. P9–11, Rhodanobacter sp. T12–5 and Trinickia sp. T12–10 inoculated alone or in combination with S. indica on tomato growth under lownutrient soil conditions. Plants were grown for 6 weeks (a) and fresh shoot weight (b) and leaf area (c) were measured. Same letters represent nonsignificantly different mean values, according to Tukey’s test (P < 0.05) after ANOVA. n = 14. Bar in the picture corresponds to 6 cm
Biocontrol of F. oxysporum and R. solani
The tomato plants treated with F. oxysporum showed typical symptoms as leaf yellowing, defoliation, stunting, wilting, necrotized xylem bundles, and death (Suppl. Fig. 4). Inoculations with the bacterial strains individually did not protect plants against the pathogen, except for strain T12–10, that significantly reduced the Fusarium disease index (Fig. 3a). Although not significantly, the magnitude of disease symptoms seemed to be mitigated when the plants were inoculated singly with the beneficial endophyte S. indica. Intriguingly, Fusarium symptoms (DI) were only significantly reduced when the plants were dual-inoculated with S. indica + P1–11, S. indica + P9–11 and S. indica + T12–10. The negative effect of Fusarium wilt on fresh weight was not significantly mitigated upon dual inoculation of S. indica + bacteria (Fig. 3b).
Effect of strains Methylobacterium sp. P1–11, Tardiphaga sp. P9–11, Rhodanobacter sp. T12–5 and Trinickia sp. T12–10 in combination or not with Serendipita indica (Si) on tomato plants against fungal pathogens. a,b) Biocontrol of Fusarium oxysporum (Fol). a Disease index (DI) score. b Fresh weight. Boxes represent standard deviation with median. Whiskers represent the Min to Max of all the values. Analysis of FW and DI was performed using the non-parametric Mann–Whitney U test (*P < 0.05, **P < 0.01, ns = non significant; n = 10); c Biocontrol of Rhizoctonia solani (Rs). Points represent mean values, and whiskers standard deviations. Same letters represent nonsignificantly different mean values, according to Tukey’s test (P < 0.05) after ANOVA (n = 3). ns = non significant
In the case of Rhizoctonia damping-off, the symptoms were first recorded 11 days after planting (dap). Single inoculation of bacteria did not restrain pathogen attack (Fig. 3c). Inoculation of seeds with S. indica typically conferred resistance against the pathogen, but the degree of plant protection in comparison to Rhizoctonia control was significant throughout the whole experiment exclusively when S. indica was co-inoculated with bacteria. More specifically, treating the plants with S. indica plus T12–10 significantly increased protection in comparison to the treatment with S. indica alone.
Effects of bacteria on root colonization by S. indica
In general, the extent of root colonization by S. indica was low, although similar results have been reported in tomato plants when S. indica was grown under the same growing conditions (Andrade-Linares et al. 2013). Ten days after planting, addition of Methylobacterium P1–11 and Trinickia T12–10 to the S. indica inoculum significantly increased the extent of root colonization in comparison to single inoculations (Figs. 4a-b). Eight weeks after planting, inoculation of S. indica with T12–10 significantly increased the number of hyphae aggregates respect to control plants (Fig. 4c). However, in all the sampled root fragments we detected the presence of the fungus and there were no differences in terms of root areas colonized by S. indica among the different treatments 8 weeks after planting (Fig. 4d). Regarding the number of spores inside the roots we observed a modest increment in plants dual-inoculated with S. indica and T12–10 (Suppl. Fig. 5). The number of spores detected in the treatment S. indica + T12–10 was significantly increased when compared to the treatment S. indica + P1–11, but not when compared to control plants. However, more hyphae producing spores were detected in S. indica + T12–10 compared to control (data not shown).
Effect of bacterial strains on root colonization by S. indica. Ten days after planting (a-b), both strains increased the root areas colonized by S. indica. Eight weeks after planting (c-d), S. indica was detected in every root fragment for every treatment (d), however the number of hyphae aggregates was significantly increased when S. indica was co-inoculated with Trinickia T12–10 (c). The boxplots show all data points, the horizontal line represents the median, surrounded by the upper (25th) and lower (75th) percentiles. The bar plots represent the means of colonization and the whiskers the standard deviations. Same letters represent non-significantly different mean values, according to Tukey’s test (P < 0.05), after ANOVA. n = 20
Stability of the mCherry-marked strains and comparison with the wild-type strain
The growth patterns and the colony and cell morphologies of the mCherry-tagged strains of Methylobacterium sp. P1–11 and Trinickia sp. T12–10 on PDA + NA w/o antibiotics were similar to those of the wild-type strains, but the colonies displayed a characteristic purple color (Suppl. Fig. 6a,b). Colonies and cells of these mCherry-marked strains were notably fluorescent under orange/red light when cultured on PDA + NA amended with kanamycin (50 μg/ml). When growing without kanamycin, few colonies of the strain Methylobacterium P1–11 (but not of Trinickia T12–10) lost the fluorescence due to plasmid loss (data not shown). The strains Tardiphaga sp. P9–11 and Rhodanobacter sp. T12–5 were not considered for further experiments as the stability of the transformation was extremely low (Supp. Figure 6c).
Physical interaction between S. indica and bacteria and root colonization patterns
To study if bacterial strains affect the morphological structure of S. indica, gfp-tagged S. indica was grown alone and in combination with mCherry-tagged bacteria. When the fungus was cultured alone, typical S. indica structures like tubular and moniliform hyphae and chlamydospores (Kost and Rexer 2013) were observed (Figs. 5a-d). During dual culture of S. indica with strains P1–11 or T12–10, fungal structures were not affected (Figs. 5e-w). Hyphae were not disrupted nor deformed, and the thickness of the hyphae remained unaltered. Intriguingly, the fungus seemed to be stimulated, and the mycelium density and the number of chlamydospores appeared to be enhanced in both co-cultures (Figs. 5e and j). Regarding their physical associations, both bacteria were spotted loosely scattered among the hyphae or attached to them (Figs. 5e-g and j-l). Sometimes bacteria aggregated around the hyphae (Figs. 5h and i), which hints at chemotaxis towards the mycelium. The strain P1–11 was often observed attached to the surface of chlamydospores, but never inside them (Figs. 5f-g). However, bacterial cells of strain T12–10 were not only found attached to the surface (Figs. 5j-m) but most strikingly, we repeatedly detected them colonizing the chlamydospores of S. indica internally (Figs. 5n-w). Some of the colonized spores were alive as a strong gfp fluorescence was detected (Figs. 5j-k and l-o) and nuclei as well as vesicles were observed (Figs. 5n-o) while others looked as empty spores with bacteria (Figs. 5j-k and p-t). Nonetheless, empty spores were also found in S. indica grown alone. Bacteria were not detected inside fungal hyphae but were further observed in living germinated spores (Suppl. Fig. 7). Moreover, in these pairings, no specialized morphological structures were observed upon colonization.
Interaction in vitro between S. indica (in green) and bacteria (in red). a Tubular hyphae and chlamydospores. b Representation of (a) in 3D. c Moniliform hyphae and ontogenies of chlamydospores. d Representation of (c) in 3D. e Bacteria loosely scattered among the hyphae and spores. f Bacteria attached to the surface of hyphae and spores, without penetrating them, and its 3D representation (g). h Bacteria attracted to the hyphae, and its 3D representation (i). j High density of chlamydospores and bacteria loosely scattered among the spores. l Bacteria attached to the spores, without penetrating them. k and m Representations of (j) and (l) in 3D. n Bacteria colonizing a spore at the germination site. o Magnification of (n) in 3D. p and s) Bacteria colonizing fungal spores internally. q and r Representations of (p) in 3D. t Transversal plane of (s) represented from the y axis and its 3D representation (u). v and w) Magnifications in 3D of the spore with arrow in (u). a, c, e-f, h, j, l, n, p, s and t: confocal images. b, d, g, i, k, m, o, q-r and u-w: 3D modeling using Imaris software
Colonization patterns of tomato plantlets by single and dual inoculations of bacteria and S. indica were investigated under sterile conditions and using marked strains. The roots of control plants (untreated) did not differ from plants inoculated with microbes, in terms of structure, cell-wall thickness and fluorescence produced by the secretion of phenolic compounds upon microbe colonization (Compant et al. 2005a, 2005b) (Figs. 6a-c). Strains P1–11 and T12–10 followed similar colonization patterns (Figs. 6d-o). Both bacteria were most abundantly detected in the upper part of the root (Figs. 6j and 7e), and considerably less abundant bacteria were found close to the root tips (Figs. 6f and n). Strain T12–10 was spotted more abundantly than P1–11 in lower parts of the roots. Analysis of colonization patterns revealed that these strains settle at the surface of the roots, and aggregate in the intercellular spaces and at lateral root emergence sites (Figs. 6d-g, 6m-n and 7b-c). They were also found internally colonizing epidermis cells (Figs. 6f-g, 5m and 7a-c) and root hairs (Figs. 6h-i, l and o). Unlike other bacteria, they were not detected at the root tip level (Compant et al. 2005a, 2005b). Very often bacteria were observed in aggregates forming biofilm-like structures on the surface of the roots (Figs. 6j-k and 7e). In the root-stem transition zone, bacteria used the stomata to gain access to the inner parts of the plants (Figs. 6d-e).
Root colonization patterns of single inoculations of S. indica (in green), Methylobacterium P1–11 (in red) and Trinickia T12–10 (in red). a and b Roots of control plants. c Root-stem transition zone with a stoma. d Bacteria colonizing the inner parts of in the root-stem transition zone through stoma (arrow) and intercellular spaces (asterisk) and a 3D representation (e). f Bacteria penetrating the exodermis cells in the lower part of the root and a 3D magnification (g). h Root hair internally colonized by bacteria and a 3D magnification (i). j Bacteria aggregating on the surface of the upper part of the root, forming biofilm-like structures (arrow). k) 3D representation of j. l Root hair colonized by bacteria. m Exodermis cell heavily colonized by bacteria in the upper part of the root, and bacteria clustering in the intercellular spaces (arrow). n Fewer colonies of bacteria penetrating intracellular spaces in the lower part of the root. o Representation in 3D of bacteria colonizing a root hair. p Hyphae loosely extended among the root hairs (asterisk) and penetrating the exodermis (arrow) in the upper part of the root and a 3D representation (q). r Loose hyphae among the root hairs in the lower part of the root and its 3D model (s). t Hyphae colonizing a root (arrow) hair and the exodermis (asterisk). A-d, f, h, j, l-n, p, r and t: confocal images. e, g, i, k, o, q, s: 3D modeling using Imaris software
Root colonization patterns of dual inoculations of S. indica (in green) with Methylobacterium P1–11 (in red) and Trinickia T12–10 (in red). a Exodermis cell heavily colonized by bacteria through intercellular spaces. b and c 3D representations of (a). d Colocalization of fungus and bacteria during colonization of root hairs (arrow), and bacteria attached to hyphae (asterisk). e Bacteria aggregating in the surface of the upper part of the root, forming biofilm-like structures. f Fungus and bacteria spotted together in lower parts of the roots. g Representation in 3D of (d). h Root hair colonized by fungus (arrow) and bacteria (asterisk). i Representation in 3D of (f). j Hyphae and bacterial aggregates colonizing the surface of the upper part of the root, and mycelium penetrating the exodermis (arrow). k Representation in 3D of (j). l Spores aggregated in the surface of the upper part of the root, surrounded by bacteria (arrow), and a 3D magnification (m). n Hyphae (arrow) and bacterial aggregates (asterisk) colonizing the surface of the root and root hairs in the lower part of the root, and its 3D representation (o). p Colocalization of bacteria, hyphae and spores (arrow) in the surface of roots close to the root tips. q 3D representation of (p). a, d-f, h, j, l, n and p: confocal images. b-c, g, I, k, m, o and q: 3D modeling using Imaris software
Mycelium of S. indica was detected along the entire length of the roots (Figs. 6p-t), but more abundantly in the upper parts. Hyphae were loosely found among the root hairs (Fig. 6p-s), attached to their surface (Figs. 7f and i) and colonizing them internally (Figs. 6t, 7d and h). Sometimes the fungus colonized in high numbers the lateral root emergence zones. The fungus also colonized the surface of the roots (Figs. 6t and 7o) as well as the interior of exodermis cells, although the hyphae displayed low fluorescence (Figs. 6p and 7j-k). No spores were detected in S. indica control plants.
When S. indica was inoculated with strain P1–11, both microbes followed the same patterns of colonization as when single inoculated (Figs. 7a-i). No differences in extent of colonization were observed. Bacteria and fungus did not exclude each other, and they were found colonizing the same areas. Bacteria were occasionally detected attached to the hyphae (Figs. 7d and g). No fungal spores were observed. Comparably, dual inoculation with S. indica and strain T12–10, resulted in similar colonization patterns as single inoculations (Figs. 7j-o). However, the area of the fungal mycelium colonization seemed to be increased in comparison to the S. indica control (data not shown). The dispersion of bacteria was increased, and they were detected more abundantly in areas close to the root tips as compared to plants single-inoculated with bacteria (Figs. 7n-o). S. indica and strain T12–10 colonized the same root zones, and bacteria were often detected adhered to hyphae (Figs. 7j-k and n-q). In contrast to plants inoculated with S. indica alone, in plants dual-inoculated we repeatedly observed spores attached to hyphae of S. indica on the surface of the roots, and these spores were often surrounded by bacteria (Figs. 7l-m and p-q).
Genome analysis
Genomic features and taxonomic affiliation
The genomes of strains Methylobacterium sp. P1–11, Tardiphaga sp. P9–11, Rhodanobacter sp. T12–5 and Trinickia sp. T12–10 were analyzed to determine which gene clusters could be linked to plant and fungal colonization. They have a total size of 6.77, 6.06, 3.96 and 6.22 Mb, and an average G + C content of 68.97%, 61.28%, 65.22% and 62.87%, respectively (Table 1). No repA genes were found in the genomes and therefore we found no evidence of plasmids in any of the strains. The taxonomical analysis only assigned species names to the strains P1–11 and T12–10, closely related to Methylobacterium sp. UNC378MF and Trinickia soli GP25–8 respectively (Suppl. Table 1). The latter strain has recently been moved to the genus Trinickia gen. Nov. (Estrada-de los Santos et al. 2018) and was previously included in the genus Paraburkholderia. Strains P9–11 and T12–5 did not reach the 95% ANI threshold required for species circumscription and were just classified at the genus level as Tardiphaga sp. and Rhodanobacter sp. respectively. The genomic features of the four genomes are summarized in Table 1.
Genes and proteins predicted to stimulate S. indica growth
Based on analogies to genes previously reported to be involved in fungal stimulation, we detected here several genes potentially involved in the stimulation of S. indica growth (Fig.8, see Suppl. Table 2 with detailed numerical values and protein annotation names). Some vitamins (e.g. B12) can solely be synthesized by bacteria (Fang et al. 2017), thus we hypothesize that vitamin production could be involved in fungal growth stimulation. We found numerous genes in all the strains implicated in the synthesis of vitamins B: thiamin (B1), riboflavin (B2), pyridoxin (B6), biotin (B7), folate (B9) and cobalamin (B12) (not in T12–5); vitamins K: menaquinone (K1) and phylloquinone (K2) as well as ubiquinone (Q10) (not in P1–11). Moreover, important genes reported to be involved in the provision of nitrogen to the fungus were identified in the four genomes, like those coding for nitrite and nitrate reductase (not in T12–5) (Zuccaro et al. 2011), ammonification and glutamine synthase, as well as the gene coding for nifU in the strains P1–11 and P9–11 (Suppl. Table 3), that is involved in nitrogen fixation (Minerdi et al. 2001). Furthermore, various genes predicted to participate in trehalose biosynthesis (Duponnois and Kisa 2006) and protein and polysaccharide secretion have been detected (Suppl. Table 2).
Circular visualization of genomes using the Blast Ring Image Generator (BRIG). Track 1: (innermost) genome map; Track 2: GC%; Track 3: GC Skew; Track 4: contig boundaries as alternating red and black color; Track 5: genome positions coding for genes potentially involved in the stimulation of fungal growth. Vitamins of the B complex (in red) are indicated by B (e.g. B12 stands for Vitamin B12); Genes involved in nitrogen supply (in black) include nitrate reductase (NR1), nitrite reductase (NR2), and nitrate/nitrite/ammonium transporters (NT). Trehalose biosynthesis (in green) is indicated by TR. Numbers (2,3,4..) before the abbreviation of genes indicated if more than one gene are present at a certain position
Genes and proteins related to plant growth promotion traits
Several genes related to plant growth promotion (PGP) features were detected in all genomes based on RAST, antiSMASH and protein functional annotation (eggNOG) analysis. The four genomes harbor genes involved in auxin biosynthesis as well as solubilization and transport of phosphates (Table 2, see Suppl. Table 4 with detailed protein and gene annotation names). Genes related to the synthesis of polyamines, phytohormone-like compounds involved in plant growth and stress mitigation (Chen et al. 2018a), were also detected. The strains Methylobacterium P1–11 and Tardiphaga P9–11 possess genes involved in nitrogen fixation (Suppl. Table 3) and genes participating in the production of nitrilases, which are implicated in the synthesis of plant hormones (Park et al. 2003). Genes involved in siderophore biosynthesis and receptors were detected; potentially contributing to the iron supply to the plant. Strains P1–11 and T12–10 encode genes for aerobactin (Table 2) and vibrioferrin (Suppl. Table 3) biosynthesis (Tanabe et al. 2003), and the antiSMASH analysis further identified biosynthetic gene clusters (BGCs) involved in the synthesis of enterobactin, pyoverdine, taiwachelin and malleobactin (Suppl. Table 5). Furthermore, strains P1–11, P9–11 and T12–10 encode the gene acdS for production of 1-aminocyclopropane-1-carboxylate (ACC) deaminase (Suppl. Table 3) related to plant growth enhancement by lowering plant ethylene levels (Hardoim et al. 2015).
Resistance to antibiotics and production of antibiotic compounds
Genes for production of antibiotic compounds as well as for antibiotic resistance may protect the plant against other pathogenic microbes. We identified several genes involved in the production of the antibiotic compounds colicin and bacteriocins (Table 2 and Suppl. Table 4). Strain Trinickia T12–10 encodes genes for type IV pili, a bacterial virulence mechanism that appears operational during pathogenesis of fungal hosts (Dörr et al. 1998), genes to produce aminoglycoside antibiotics and a cyclic lipopeptide identified as bananamide (Nguyen et al. 2017) (Suppl. Table 5). Besides, the four genomes encode proteins involved in the synthesis of phenazines (Suppl. Table 3) (Thomashow and Weller 1988), as well as BGCs identified as lipopolysaccharide (LPS) and O-antigen (glycan polymer contained within an LPS) (Suppl. Table 5). Furthermore, CAZy analysis detected enzymes involved in the breakdown of fungal cell-wall components like β-hexosaminidases, exo-β-glucosaminidase and chitooligosaccharide deacetylases (Table 4, see Suppl. Table 6 for detailed information), that have been described to degrade chitin, chitosan and chitooligosaccharides (Mekasha et al. 2016; Katta et al. 2013; Zhao et al. 2010). The RAST analysis further characterized in the genomes of T12–5 and T12–10 several genes under the subsystem “Chitin and N-acetylglucosamine utilization” (data not shown). In a similar way, the four genomes contain genes involved in multidrug resistance efflux pumps, resistance to the bactericide fluoroquinolone and synthesis of β-lactamases, enzymes that confer resistance to β-lactam antibiotics (Berg et al. 2010) (Table 2 and Suppl. Table 4).
Genes and proteins related to stress tolerance
Numerous genes identified in these genomes participate in multiple pathways of protection against environmental stress (Table 2 and Suppl. Tables 4 and 5), and this protection may ultimately result in plant growth promotion (Liu et al. 2017). For instance, genes involved in cell protection against oxidative stress, like those for the synthesis of peroxidases, superoxide dismutases, hydroperoxide reductases, catalases, glutathione S-transferases and carotenoids (Suppl. Table 5). They further harbor genes for detoxification, like those coding for cyanide hydratases/nitrilases (Howden and Preston 2009) and rhodanese (Chaudhary and Gupta 2012).
In addition, the four strains might enhance plant resistance to salt stress by producing the osmoregulator glycine betaine (Table 2 and Suppl. Table 4) (Ashraf and Foolad 2007) and the disaccharide trehalose (Penna 2003), as well as genes that operate directly in the protection of membranes and proteins, and functionally in plant immunity, like cold and heat shock proteins, chaperones and glucans (Park and Seo 2015). Moreover, these strains contain numerous copies of genes implicated in plant resistance to heavy metals/metalloids including arsenic, mercury, chromium, copper, cadmium, cobalt and zinc (Table 2 and Suppl. Table 4).
Genes implicated in colonization and establishment of endophytes in the host
Induction of flagellar activity and chemotaxis (Pinski et al. 2019) and attachment to the root surface are decisive steps for bacteria to colonize roots (Mitter et al. 2013). It has been reported that bacterial lipopolysaccharides (LPS), exopolysaccharides (EPS), Type IV pili and secretion systems are involved in attachment and colonization (Balsanelli et al. 2010; Meneses et al. 2011; Dörr et al. 1998; Sessitsch et al. 2012). The four bacteria encode numerous genes involved in flagellar activity, chemotaxis, secretion systems and LPS assembly (Table 3, Suppl. Table 5). Strain Methylobacterium P1–11 has also genes for EPS biosynthesis (Suppl. Table 2) and strains Rhodanobacter T12–5 and Trinickia T12–10 have genes involved in Type IV pili.
Cell-wall degrading enzymes are important traits to gain access to the intracellular spaces (Compant et al. 2010). We identified numerous genes related to plant cell wall-degrading enzymes (PCWDEs) within the CAZy families carbohydrate esterases (CE) and glycoside hydrolases (GH), with the capabilities of degrading cellulose, hemicellulose and pectin (Table 4). The RAST analysis further detected genes implicated in lignin catabolism in strains P1–11, P9–11 and most notably T12–10 (results not shown). Many other CAZy enzymes related to the breakdown of common plant intracellular poly- and oligosaccharides were detected (Table 4 and Suppl. Table 3).
Interestingly, strains P1–11, P9–11 and T12–10 encode the gene oxc implicated in the catabolism of oxalate (Table 2), which has been reported to be required for successful plant colonization by the endophyte Paraburkholderia phytofirmans PsJN (Kost et al. 2014). Furthermore, these strains harbor numerous genes for ROS-detoxification (Table 2), required for overcoming the plant defense response upon colonization (Kandel et al. 2017). All these genomic features presumably permit these bacteria to colonize roots and adapt to the endophytic lifestyle.
Discussion
The interactions between fungi and bacteria have often been overlooked, but they seem to be more widespread than expected and their dynamics may be crucial in natural ecosystems (Deveau et al. 2018). Here, these endophytic bacteria are suspected to positively interact with the functioning of the fungal symbiosis, therefore they might be considered as mycorrhizal helper bacteria (Frey-Klett et al. 2007). These strains greatly promoted fungal growth in vitro, enhanced plant growth, and their combination with S. indica could effectively confer resistance against plant pathogens. We previously found other strains within the genus Mycolicibacterium exhibiting similar traits although the functioning underlying the tripartite interactions seem to be partly distinct.
Effective microbial inoculum must colonize plant roots and overcome the plant immune system (Compant et al. 2005a; Liu et al. 2017). In the genomes of the tested bacterial strains the presence of multiple genes typically considered to be involved in root colonization and endophytism was congruent with microscopy observations in which root exodermis cells and hairs were internally colonized by strains Methylobacterium P1–11 and Trinickia T12–10. These strains were detected in different root zones, and particularly strain T12–10 expanded more throughout the root, probably due to the higher number of genes involved in flagellar motility and secretion systems. In addition to the bacterial genomic features, it has been reported that plants might also facilitate the entry of bacteria into periplasmic spaces in root cells, so that plants might acquire nutrients from bacterial protoplasts in a process known as rhizophagy cycle (White et al. 2018). In the process of root colonization S. indica and bacteria did not exclude each other but they were commonly co-localized, and rather than competing they seem to cooperate, since the addition of bacteria resulted in increased hyphae expansion, fungal sporulation and root zones colonized by the fungus. Interestingly, we often found bacteria attached to fungal hyphae, hence bacteria might employ hyphae as vectors to colonize new niches along the root (Kohlmeier et al. 2005; Miquel Guennoc et al. 2018).
Most striking was to find cells of Trinickia T12–10 colonizing spores of S. indica, thus the endosymbiotic associations among Sebacinales and bacteria could be much broader in nature than we previously thought. Congruently with this finding, bacteria from the order Burkholderiales are among the most frequently identified intracellular bacteria in fungal hyphae (Partida-Martinez and Hertweck 2005; Frey-Klett et al. 2011) and several Burkholderia sensu lato strains are endosymbionts of beneficial endophytic fungi (Compant et al. 2008). Moebius et al. (2014) revealed that a type 2 secretion system (T2SS) of Burkholderia rhizoxinica is required for the endosymbiosis with Rhizopus microsporus. In that study, mutants deficient in the genes gspC and gspD (general secretion pathways) were unable to internally colonize the fungus and induce sporulation. Among these bacteria, strain T12–10 contains the highest number of genes involved in T2SS, including gspC and gspD, which presumably enables this strain to adhere and colonize fungal structures. Strain P1–11, however, lacks these genes and harbors fewer T2SS genes. This could explain why T12–10 could internally colonize the spores and induce sporulation, while P1–11 did not enter spores. Strain T12–10 encodes also genes involved in fungal cell wall degradation (e.g. β-hexosaminidase), although it lacks the gene (chi) for chitinase production, which was shown to be crucial for entering fungal hyphae in the B. rhizoxinica-R. microsporus symbiosis. Consequently, it is not yet understood how Trinickia T12–10 gains access to the spores of S. indica. Likewise, the mechanism by which RrF4 invades S. indica is yet unresolved. This bacterium lacks the chi gene and even T2SS genes, albeit concomitant with T12–10, genes for T4SS, involved in adhesion, are present (Glaeser et al. 2016). It has been hypothesized that germinating spores of S. indica can be potential entry sites for bacteria (Guo et al. 2017), and for penetrating growing hyphal tips where walls are thin, other enzymes different from chitinases (e.g. glucanases) might suffice. We detected T12–10 in apparently empty spores, but also inside those with an intact cytoplasm as well germinated spores. Nevertheless, it claims further investigation if the bacterium T12–10 could be vertically transmitted to the next generation of spores.
The rhizosphere and the roots of most plants are host of complex interactions between different microorganisms, but the precise nature and mechanisms of these interactions are yet to be elucidated. To obtain further understanding on potential mechanisms, we analyzed the genomes of these bacterial helpers. We detected a great number of genes related to vitamin production presumably involved in fungal growth stimulation. For instance, cobalamine (vitamin B12) produced by bacteria was reported to be implicated in the promotion of fungal growth and fitness (Ghignone et al. 2012). Jiang et al. (2018) found that S. indica is auxothophic for thiamine (vitamin B1), and the bacterium Bacillus subtilis complemented S. indica’s auxotrophy for thiamine and promoted its growth. S. indica holds biotroph-associated genomic adaptations, hence some genes and protein families have undergone contraction, like those for nitrate and nitrite reductase, amino acid and ABC transporters, and is thus predicted to suffer from some metabolic deficiencies. Congruently, the fungus barely grows on nitrate as sole N source (Zuccaro et al. 2011). The genomes of the bacterial strains tested in this study harbor several genes coding for nitrate and nitrate reductase, amino acid and peptide ABC transporters, ammonium transporters and glutamine synthase as well as nitrogen fixation, which might enhance fungal nitrogen metabolism, palliating the predicted deficiencies. Furthermore, Salvioli et al. (2016) revealed that an endobacterium of Gigaspora margarita enhanced fungal sporulation and increased the bioenergetic capacity of the mycorrhizal fungus by inducing oxidative phosphorylation and ATP production. Investigating S. indica, Bhuyan et al. (2015) detected that genes coding for enolase-I and urease D, which are involved in the activation of several hexose sugar transporters and in the glycolytic pathway for generating ATP, were up-regulated in the presence of the fungal-stimulating bacterium Azotobacter chroococcum WR5. Similarly, the bacteria tested in this study, and particularly strain T12–10 that can internally colonize fungal structures, might up-regulate sugar uptake, glycolysis and oxidative phosphorylation, prompting ATP production and increased fungal fitness. There is still no clear evidence if only one, or several genomic features detected in these bacteria are responsible for fungal growth stimulation. Nevertheless, these isolates did not stimulate the growth of the fungal pathogens, as opposed to S. indica. Hence there seems to exist specificity in the stimulation of fungal growth, as it has been earlier reported for other mycorrhizal helper bacteria (Frey-Klett et al. 2007). To better elucidate the molecular basis behind the fungal stimulation, further studies should focus on the functional role of these bacterial factors by knock-out mutants, and transcriptomics of fungus and bacteria to detect genes up and down-regulated during co-cultivation.
The tested bacteria harbor also many genes involved in plant growth promotion, which confirms the potential of these strains as plant biostimulants. Correspondingly, strains Tardiphaga P9–11, Rhodanobacter T12–5 and Trinickia T12–10 increased plant fresh weight and leaf area, although strain Methylobacterium P1–11 did not affect plant growth. In agreement with the widely reported beneficial effect of S. indica on plant growth (Franken 2012), S. indica significantly increased tomato growth in this experiment. Dual inoculations, however, did not further improve the growth-promoting effect exerted by microbes inoculated singly. Interestingly, inoculation of S. indica + T12–5 significantly displayed lower performance than single inoculations. Certainly, there are several reports of combinations of fungi and bacteria providing no better or, in some cases, worse plant promotion than the isolates used singly (Whipps 2001). In our previous study, combinations of S. indica with strain P9–22 or P9–64 of Mycolicibacterium negatively affected plant growth in comparison to single inoculations (del Barrio-Duque et al. 2019). The same negative effect was described for co-inoculation of pseudomonad R62 and S. indica (Sarma et al. 2011). This incompatibility of fungal-bacterial inoculants has been ascribed to competition for nutrients and niches in the rhizosphere (Whipps 2001), although in this study S. indica and bacteria seemed to cooperate during colonization. The lack of synergism might also be caused by alterations in the plant hormone levels. Rather low levels of exogenous auxins stimulate plant growth whereas high concentration may cause a negative impact (Sarwar and Frankenberger 1994). Since these bacteria and S. indica can produce auxins (Sirrenberg et al. 2007), the dual inoculation might lead to an imbalance of auxins levels and consequently less growth promotion. In analogy, a further explanation may be over-colonization of roots by the fungus. S. indica begins colonization with a biotrophic growth phase invading living cells, followed by a cell death-dependent phase, in which root cells are actively killed by the fungus (Qiang et al. 2012b). If the number of dead cells transcends a threshold, S. indica could manifest a negative influence on plant growth (Fakhro et al. 2010). Our isolates greatly stimulated fungal growth in vitro, and by microscopy observations we detected enhanced hyphae expansion in plants dual-inoculated with S. indica and strain T12–10. Bacteria might stimulate fungal growth and root colonization until the threshold is surpassed, shifting the fungus thus from mutualistic to saprotrophic and impairing plant development.
On the basis of the accelerated depletion of the currently available fungicides, and the huge amounts of crops affected worldwide by plant pathogens, the upcoming generation of bioinoculants shall combine plant growth enhancement with biological control of plant pathogens (Collinge et al. 2019). Notwithstanding that these bacterial strains harbor genes involved in biocontrol of plant diseases, bacteria inoculated individually were generally not able to control fungal pathogens. This fact complies with the in vitro tests in which confrontations between bacteria and the pathogens did not display a direct suppression of fungal growth. Only the strain Trinickia T12–10 significantly reduced the progression and severity of Fusarium wilt. In agreement, numerous strains of Burkholderia sensu lato have been described to induce plant resistance against fungal pathogens (Compant et al. 2008).
It has been reported that S. indica colonization results in the activation of defense related genes and ISR, which consequently improves plant tolerance against biotic stresses (reviewed in Franken 2012; Gill et al. 2016). Consistently, in this study S. indica also reduced disease progression caused by fungal pathogens. However, the highest degree of protection was observed when the plants were dual-inoculated with combinations of S. indica and bacteria. This synergistic effect was also observed in our previous research, in which Fusarium wilt’s severity was significantly reduced by dual-inoculations of S. indica with Mycolicibacterium strains P1–18 and P9–22 in comparison to single inoculations, and biocontrol of Rhizoctonia resulted most effective by combining S. indica with Mycolicibacterium P9–64 (del Barrio-Duque et al. 2019). Interestingly, the endofungal bacterium of S. indica (Rhizobium radiobacter F4) was found to induce resistance responses in barley and confer resistance against different pathogens (Sharma et al. 2008; Glaeser et al. 2016). Consequently, it was speculated that the beneficial effect exerted by this fungus might actually be attributable to the bacterium (Qiang et al. 2012a; Glaeser et al. 2016). Following this line, it is likely that other bacterial strains might also contribute to further increase the beneficial effect reported for this fungus. There might be a synergism in rising the plant ISR (defense) when S. indica and certain bacteria are inoculated. Genome analysis of the bacterial strains tested in this study revealed several genes involved in the activation of the plant ISR, like those involved in the synthesis of PCWDEs, phenazines, LPS and O-antigens, type IV pili, antibiotics, EPS and flagella (Pieterse et al. 2014; Van Loon et al. 1998; Lerouge and Vanderleyden 2002) that might complement the S. indica-triggered ISR. In analogy, there might be a synergism in increasing antioxidant activity in plant cells. S. indica induces systemic resistance by increasing the concentration of antioxidants, glutathione and ascorbate in the plants to mitigate the oxidative stress caused by pathogens (Waller et al. 2005; Gill et al. 2016). Since our bacterial strains encode numerous genes involved in the resistance to oxidative stress, like glutathione S-transferase, peroxidase, and genes participating in the synthesis of carotenoids (Stahl and Sies 2003), we can hypothesize that with the addition of bacterial helpers to the fungal inoculum, the plant has additional “tools” to cope with the oxidative stress caused by pathogens. Besides, on the basis of ethylene as a stress hormone, potential production of ACC-deaminase by these bacteria might have an additional asset to protect plants (Wang et al. 2000). Furthermore, the genomes of the tested bacteria encode also genes involved in direct biocontrol of pathogens, like genes involved in siderophores synthesis and receptors that deprive the pathogens from iron, enzymes involved in the degradation of fungal cell-wall structures, phenazines or cyclic lipopeptides (Whipps 2001; Compant et al. 2005a). The outcome of the multipartite interaction among these beneficial microbes, the pathogens and the plant might rely on several of these mechanisms. Perhaps the combined effect of competition for nutrients and space, S. indica-triggered ISR and the bacterial features directly and indirectly involved in biocontrol might justify the increased resistance of plants inoculated with S. indica + bacteria.
All in all, what really happens inside the roots and the rhizosphere seems to be a complex network of multitrophic cooperation and several factors drive the outcome of the plant-microbe interaction. When searching for plant growth enhancement, the application of beneficial microbes should consider the nutrient conditions of the soil. In this study, dual and single inoculations of fungus and bacteria in rich soil did not promote shoot growth while on low-nutrient soil it did. This nutrient-dependent effect upon S. indica inoculation on tomato was previously reported (Fakhro et al. 2010). This research group also demonstrated that the concentration of S. indica inoculum was crucial, and negative effects were observed when high cell doses were applied. Similarly, inoculation of S. indica with a high concentration of its endobacterium resulted in harmful, if not lethal, effects to the fungus (Sharma et al. 2008). It is evident that the optimal concentration of S. indica, and the bacteria to accompany the fungal inoculum, should be further investigated and additional research is needed to further elucidate the interaction of the growth-promoting fungus and its bacterial partners. Soil condition, plant determinants (physiology, species and cultivar), environmental factors, dosage of the inoculum and microbial compatibility must be addressed for the development of new biostimulants and biocontrol agents.
References
Alikhan NF, Petty NK, Zakour NLB, Beatson SA (2011) BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. https://doi.org/10.1186/1471-2164-12-402
Andrade-Linares DR, Müller A, Fakhro A, Schwarz D, Franken P (2013) Impact of Piriformospora indica on tomato. In: Varma A et al (eds) Piriformospora indica, Soil biology, vol 33. Springer-Verlag, Berlin, Heidelberg, pp 107–117
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59:206–216. https://doi.org/10.1016/j.envexpbot.2005.12.006
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Balsanelli E, Serrato RV, De Baura VA, Sassaki G et al (2010) Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environ Microbiol 12:2233–2244. https://doi.org/10.1111/j.1462-2920.2010.02187.x
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Berg G (2009) Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol 84:11–18. https://doi.org/10.1007/s00253-009-2092-7
Berg G, Egamberdieva D, Lugtenberg B, Hagemann M (2010) Symbiotic plant–microbe interactions: stress protection, plant growth promotion, and biocontrol by Stenotrophomonas. In: Seckbach J et al (eds) Symbioses and stress, Cellular origin, life in extreme habitats and astrobiology, vol 17. Springer, Dordrecht, pp 445–460
Bhuyan SK, Bandyopadhyay P, Kumar P, Mishra DK, Prasad R et al (2015) Interaction of Piriformospora indica with Azotobacter chroococcum. Sci Rep 5:13911. https://doi.org/10.1038/srep13911
Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy HV, Bonfante P (1996) An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl Environ Microbiol 62:3005–3010
Bonfante P, Anca IA (2009) Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol 63:363–383. https://doi.org/10.1146/annurev.micro.091208.073504
Cardinale M, Berg G (2015) Visualization of plant-microbe interactions. In: Lugtenberg B (ed) Principles of plant-microbe interactions. Springer, Cham, pp 299–306
Chaudhary M, Gupta R (2012) Cyanide detoxifying enzyme: rhodanese. Curr Biotech 1:327–335. https://doi.org/10.2174/2211550111201040327
Chaumeil P. A, Mussig A. J, Hugenholtz P, Parks D. H (2019) GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics btz848. https://doi.org/10.1093/bioinformatics/btz848
Chen D, Shao Q, Yin L, Younis A, Zheng B (2018a) Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front Plant Sci 9. https://doi.org/10.3389/fpls.2018.01945
Chen S, Zhou Y, Chen Y, Gu J (2018b) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:884–i890. https://doi.org/10.1093/bioinformatics/bty560
Collinge D. B, Jørgensen H. J. L, Latz M, Manzotti A, Ntana F, Rojas Tayo E. C, Jensen B (2019) Searching for novel fungal biological control agents for plant disease control among endophytes. In: Hodkinson T et al (eds) Endophytes for a Growing World. Cambridge University Press, 25–51. https://doi.org/10.1017/9781108607667.003
Compant S, Duffy B, Nowak J, Clément C, Barka EA (2005a) Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol 71:4951–4959. https://doi.org/10.1128/AEM.71.9.4951-4959.2005
Compant S, Reiter B, Sessitsch A, Nowak J, Clément C, Barka EA (2005b) Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol 71:1685–1693. https://doi.org/10.1128/AEM.71.4.1685-1693.2005
Compant S, Nowak J, Coenye T, Clément C, Ait Barka E (2008) Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev 32:607–626. https://doi.org/10.1111/j.1574-6976.2008.00113.x
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. https://doi.org/10.1016/j.soilbio.2009.11.024
Constantin ME, de Lamo FJ, Rep M, Takken FL (2019) Endophyte-mediated resistance in tomato to Fusarium oxysporum is independent of ET, JA and SA. Front Plant Sci 10:979. https://doi.org/10.3389/fpls.2019.00979
De Clercq D, Van Trappen S, Cleenwerck I, Ceustermans A, Swings J et al (2006) Rhodanobacter spathiphylli sp. nov., a gamma proteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. IJSEM 56:1755–1759. https://doi.org/10.1099/ijs.0.64131-0
de Lamo FJ, Constantin ME, Fresno DH, Boeren S, Rep M, Takken FL (2018) Xylem sap proteomics reveals distinct differences between R gene-and endophyte-mediated resistance against Fusarium wilt disease in tomato. Front Microbiol 9:2977. https://doi.org/10.3389/fmicb.2018.02977
del Barrio-Duque A, Ley J, Samad A, Antonielli L, Sessitsch A, Compant S (2019) Beneficial endophytic bacteria-Serendipita indica interaction for crop enhancement and resistance to phytopathogens. Front Microbiol 10:2888. https://doi.org/10.3389/fmicb.2019.02888
Delignette-Muller M. L, Dutang C (2015) Fitdistrplus: an R package for fitting distributions. J Stat Softw. 64:1-34. https://doi.org/10.18637/jss.v064.i04
Deshmukh S, Hückelhoven R, Schäfer P, Imani J, Sharma M, Weiss M, Kogel KH (2006) The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. PNAS 103:18450–18457. https://doi.org/10.1073/pnas.0605697103
Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, Hacquard S, Hervé V, Labbé J, Lastovetsky OA, Mieszkin S, Millet LJ, Vajna B, Junier P, Bonfante P, Krom BP, Olsson S, van Elsas JD, Wick LY (2018) Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev 42:335–352. https://doi.org/10.1093/femsre/fuy008
Di Pietro A, Roncero MIG (1996) Endopolygalacturonase from Fusarium oxysporum f sp lycopersici: purification, characterization, and production during infection of tomato plants. Phytopathology 86:1324–1330
Dörr J, Hurek T, Reinhold-Hurek B (1998) Type IV pili are involved in plant–microbe and fungus–microbe interactions. Mol 30:7–17. https://doi.org/10.1046/j.1365-2958.1998.01010.x
Duponnois R, Kisa M (2006) The possible role of trehalose in the mycorrhiza helper bacterium effect. Botany 84:1005–1008. https://doi.org/10.1139/b06-053
Estrada-de los Santos, P, Palmer, M, Chávez-Ramírez et al (2018) Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9:389. https://doi.org/10.3390/genes9080389
Fakhro A, Andrade-Linares DR, von Bargen S, Bandte M, Büttner C, Grosch R, Schwarz D, Franken P (2010) Impact of Piriformospora indica on tomato growth and on interaction with fungal and viral pathogens. Mycorrhiza 20:191–200. https://doi.org/10.1007/s00572-009-0279-5
Fang H, Kang J, Zhang D (2017) Microbial production of vitamin B 12: a review and future perspectives. Microb Cell Factories 16:15. https://doi.org/10.1186/s12934-017-0631-yFastQC. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Franken P (2012) The plant strengthening root endophyte Piriformospora indica: potential application and the biology behind. Appl Microbiol Biotechnol 96:1455–1464. https://doi.org/10.1007/s00253-012-4506-1
Frey-Klett P, Garbaye JA, Tarkka M (2007) The mycorrhiza helper bacteria revisited. New Phytol 176:22–36. https://doi.org/10.1111/j.1469-8137.2007.02191.x
Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A (2011) Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75:583–609. https://doi.org/10.1128/MMBR.00020-11
Galata V, Fehlmann T, Backes C, Keller A (2018) PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res 47:D195–D202. https://doi.org/10.1093/nar/gky1050
Garbaye J (1994) Helper bacteria: a new dimension to the mycorrhizal symbiosis (Tansley review, 76). New Phytol 128:197–210
Gawehns F, Houterman PM, Ichou FA, Michielse CB, Hijdra M, Cornelissen BJC et al (2014) The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol plant-microbe interact 27:336–348. https://doi.org/10.1094/MPMI-11-13-0330-R
Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L, Cruveiller S, Bianciotto V, Piffanelli P, Lanfranco L, Bonfante P (2012) The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J 6:136–145. https://doi.org/10.1038/ismej.2011.110
Gill SS, Gill R, Trivedi DK, Anjum NA, Sharma KK, Ansari MW, Ansari AA, Johri AK, Prasad R, Pereira E, Varma A, Tuteja N (2016) Piriformospora indica: potential and significance in plant stress tolerance. Front Microbiol 7:332. https://doi.org/10.3389/fmicb.2016.00332
Glaeser SP, Imani J, Alabid I, Guo H, Kumar N, Kämpfer P, Hardt M, Blom J, Goesmann A, Rothballer M, Hartmann A, Kogel KH (2016) Non-pathogenic Rhizobium radiobacter F4 deploys plant beneficial activity independent of its host Piriformospora indica. ISME J 10:871–884. https://doi.org/10.1038/ismej.2015.163
Glassner H, Zchori-Fein E, Compant S, Sessitsch A, Katzir N, Portnoy V, Yaron S (2015) Characterization of endophytic bacteria from cucurbit fruits with potential benefits to agriculture in melons (Cucumis melo L). FEMS Microbiol Ecol 91:7. https://doi.org/10.1093/femsec/fiv074
Guetsky R, Shtienberg D, Elad Y, Dinoor A (2001) Combining biocontrol agents to reduce the variability of biological control. Phytopathology 91:621–627. https://doi.org/10.1094/PHYTO.2001.91.7.621
Guo H, Glaeser SP, Alabid I, Imani J, Haghighi H, Kämpfer P, Kogel KH (2017) The abundance of endofungal bacterium Rhizobium radiobacter (syn Agrobacterium tumefaciens) increases in its fungal host Piriformospora indica during the tripartite sebacinalean symbiosis with higher plants. Front Microbiol 8:629. https://doi.org/10.3389/fmicb.2017.00629
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A et al (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320. https://doi.org/10.1128/MMBR.00050-14
Hervás A, Landa B, Datnoff LE, Jiménez-Díaz RM (1998) Effects of commercial and indigenous microorganisms on Fusarium wilt development in chickpea. Biol Control 13:66–176. https://doi.org/10.1006/bcon.1998.0659
Hoffman MT, Arnold AE (2010) Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol 76:4063–4075. https://doi.org/10.1128/AEM.02928-09
Howden AJ, Preston GM (2009) Nitrilase enzymes and their role in plant–microbe interactions. Microbial Biotechnol 2:441–451. https://doi.org/10.1111/j.1751-7915.2009.00111.x
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P (2015) eggNOG 45: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. https://doi.org/10.1093/nar/gkv1248
Jiang X, Zerfaß C, Feng S, Eichmann R, Asally M, Schäfer P, Soyer OS (2018) Impact of spatial organization on a novel auxotrophic interaction among soil microbes. The ISME journal 12(6):1443–1456. https://doi.org/10.1038/s41396-018-0095-z
Kandel S, Joubert P, Doty S (2017) Bacterial endophyte colonization and distribution within plants. Microorganisms 5:77. https://doi.org/10.3390/microorganisms5040077
Katta S, Ankati S, Podile AR (2013) Chitooligosaccharides are converted to N-acetylglucosamine by N-acetyl-β-hexosaminidase from Stenotrophomonas maltophilia. FEMS Microbiol Lett 348:19–25. https://doi.org/10.1111/1574-6968.12237
Kohlmeier S, Smits TH, Ford RM, Keel C, Harms H, Wick LY (2005) Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol 39:4640–4646. https://doi.org/10.1021/es047979z
Kost G, Rexer KH (2013) Morphology and ultrastructure of Piriformospora indica. In: Varma A et al (eds) Piriformospora indica, Soil biology, vol 33. Springer-Verlag, Berlin, Heidelberg, pp 25–36
Kost T, Stopnisek N, Agnoli K, Eberl L, Weisskopf L (2014) Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front Microbiol 4:421. https://doi.org/10.3389/fmicb.2013.00421
Kumar V, Sahai V, Bisaria VS (2011) High-density spore production of Piriformospora indica, a plant growth-promoting endophyte, by optimization of nutritional and cultural parameters. Bioresour Technol 102:3169–3175. https://doi.org/10.1016/j.biortech.2010.10.116
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nature Meth 9:357–359
Lenth R (2019) Emmeans: estimated marginal means, aka least-squares means. R package version 1.3.5.1. https://CRAN.R-project.org/package=emmeans
Lerouge I, Vanderleyden J (2002) O-antigen structural variation: mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol Rev 26:17–47. https://doi.org/10.1111/j.1574-6976.2002.tb00597.x
Liu H, Carvalhais LC, Crawford M, Singh E, Dennis PG, Pieterse CM, Schenk PM (2017) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front Microbiol 8:2552. https://doi.org/10.3389/fmicb.2017.02552
Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B (2013) The carbohydrate-active enzymes database (CAZy). Nucleic Acids Res 42:D490–D4D5. https://doi.org/10.1093/nar/gkt1178
Martínez-Arias C, Macaya-Sanz D, Witzell J, Martín JA (2019) Enhancement of Populus alba tolerance to Venturia tremulae upon inoculation with endophytes showing in vitro biocontrol potential. Eur J Plant Pathol 153:1031–1042. https://doi.org/10.1007/s10658-018-01618-6
Meena KK, Mesapogu S, Kumar M, Yandigeri MS, Singh G, Saxena AK (2010) Co-inoculation of the endophytic fungus Piriformospora indica with the phosphate-solubilising bacterium Pseudomonas striata affects population dynamics and plant growth in chickpea. Biol Fert Soils 46:169–174. https://doi.org/10.1007/s00374-009-0421-8
Mekasha S, Toupalová H, Linggadjaja E, Tolani HA, Anděra L, Arntzen MØ, Vaaje-Kolstad G, Eijsink VGH, Agger JW (2016) A novel analytical method for d-glucosamine quantification and its application in the analysis of chitosan degradation by a minimal enzyme cocktail. Carbohydr Res 433:18–24. https://doi.org/10.1016/j.carres.2016.07.003
Meneses CH, Rouws LF, Simões-Araújo JL, Vidal MS, Baldani JI (2011) Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol Plant-Microbe In 24:1448–1458. https://doi.org/10.1094/MPMI-05-11-0127
Minerdi D, Fani R, Gallo R, Boarino A, Bonfante P (2001) Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl Environ Microbiol 67:725–732. https://doi.org/10.1128/AEM.67.2.725-732.2001
Miquel Guennoc C, Rose C, Labb, J, Deveau, A (2018) Bacterial biofilm formation on the hyphae of ectomycorrhizal fungi: a widespread ability under controls? FEMS Microbiol Ecol 94:fiy093. https://doi.org/10.1093/femsec/fiy093
Mitter B, Petric A, Shin MW, Chain PS, Hauberg-Lotte L et al (2013) Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci 4:120. https://doi.org/10.3389/fpls.2013.00120
Moebius N, Üzüm Z, Dijksterhuis J, Lackner G, Hertweck C (2014) Active invasion of bacteria into living fungal cells. Elife 3:e03007. https://doi.org/10.7554/eLife.03007.001
Mosse B (1970) Honey-coloured, sessile Endogone spores: II. Changes in fine structure during spore development. Arch. Microbiol.74:146. https://doi.org/10.1007/BF00446902
Nautiyal CS, Chauhan PS, DasGupta SM, Seem K, Varma A, Staddon WJ (2010) Tripartite interactions among Paenibacillus lentimorbus NRRL B-30488, Piriformospora indica DSM 11827, and Cicer arietinum L. World J Microbiol Biotechnol 26:1393–1399. https://doi.org/10.1007/s11274-010-0312-z
Nguyen DD, Melnik AV, Koyama N, Lu X, Schorn M, Fang J, Aguinaldo K, Lincecum TL Jr, Ghequire MGK, Carrion VJ, Cheng TL, Duggan BM, Malone JG, Mauchline TH, Sanchez LM, Kilpatrick AM, Raaijmakers JM, de Mot R, Moore BS, Medema MH, Dorrestein PC (2017) Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat Microbiol 2:16197. https://doi.org/10.1038/nmicrobiol.2016.197
Okonechnikov K, Conesa A, García-Alcalde F (2015) Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294. https://doi.org/10.1093/bioinformatics/btv566
Park CJ, Seo YS (2015) Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol J 31:323–333. https://doi.org/10.5423/PPJ.RW.08.2015.0150
Park WJ, Kriechbaumer V, Müller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E (2003) The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol 133:794–802. https://doi.org/10.1104/pp.103.026609
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. https://doi.org/10.1101/gr.186072.114
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. https://doi.org/10.1038/nbt.4229
Partida-Martinez LP, Hertweck C (2005) Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888. https://doi.org/10.1038/nature03997
Penna S (2003) Building stress tolerance through over-producing trehalose in transgenic plants. Trends Plant Sci 8:355–357. https://doi.org/10.1016/S1360-1385(03)00159-6
Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375
Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team (2019) nlme: Linear and Nonlinear Mixed Effects Models. R package version 31–140. https://CRAN.R-project.org/package=nlme
Pinski A, Betekhtin A, Hupert-Kocurek K, Mur LA, Hasterok R (2019) Defining the genetic basis of plant–endophytic bacteria interactions. Int J Mol Sci 20:1947. https://doi.org/10.3390/ijms20081947
Pontecorvo G, Roper JA, Chemmons LM, MacDonald KD, Bufton AWJ (1953) The genetics of Aspergillus nidulans. Adv Genet 5:141–238. https://doi.org/10.1016/S0065-2660(08)60408-3
Qiang X, Weiss M, Kogel KH, Schäfer P (2012a) Piriformospora indica—a mutualistic basidiomycete with an exceptionally large plant host range. Mol Plant Pathol 13:508–518. https://doi.org/10.1111/j.1364-3703.2011.00764.x
Qiang X, Zechmann B, Reitz MU, Kogel KH, Schäfer P (2012b) The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress–triggered caspase-dependent cell death. Plant Cell 24:794–809. https://doi.org/10.1105/tpc.111.093260
Salvioli A, Ghignone S, Novero M, Navazio L, Bagnaresi P, Bonfante P (2016) Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J 10:130–144. https://doi.org/10.1038/ismej.2015.91
Sarma MVRK, Kumar V, Saharan K, Srivastava R, Sharma AK, Prakash A, Sahai V, Bisaria VS (2011) Application of inorganic carrier-based formulations of fluorescent pseudomonads and Piriformospora indica on tomato plants and evaluation of their efficacy. J Appl Microbiol 111:456–466. https://doi.org/10.1111/j.1365-2672.2011.05062.x
Sarwar M, Frankenberger WT (1994) Influence of L-tryptophan and auxins applied to the rhizosphere on the vegetative growth of Zea mays L. Plant Soil 160:97–104. https://doi.org/10.1007/BF00150350
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Sessitsch A, Coenye T, Sturz AV, Vandamme P, Barka EA, Salles JF et al (2005) Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int J Syst Evol Microbiol 55:1187–1192. https://doi.org/10.1099/ijs.0.63149-0
Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, van Overbeek L, Brar D, van Elsas JD, Reinhold-Hurek B (2012) Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant-Microbe In 25:28–36. https://doi.org/10.1094/MPMI-08-11-0204
Sharma M, Schmid M, Rothballer M, Hause G, Zuccaro A, Imani J, Kämpfer P, Domann E, Schäfer P, Hartmann A, Kogel KH (2008) Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol 10:2235–2246. https://doi.org/10.1111/j1462-5822200801202x
Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, de Lorenzo V (2013) The standard European vector architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666–D675. https://doi.org/10.1093/nar/gks1119
Simon A, Bindschedler S, Job D, Wick LY, Filippidou S et al (2015) Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol Ecol 91:fiv116. https://doi.org/10.1111/j.1574-6941.2007.00425.x
Sirrenberg A, Göbel C, Grond S, Czempinski N, Ratzinger A, Karlovsky P, Santos P, Feussner I, Pawlowski K (2007) Piriformospora indica affects plant growth by auxin production. Physiol Plant 131:581–589. https://doi.org/10.1111/j1399-3054200700983x
Stahl W, Sies H (2003) Antioxidant activity of carotenoids. Mol Asp Med 24:345–351. https://doi.org/10.1016/S0098-2997(03)00030-X
Svenningsen NB, Watts-Williams SJ, Joner EJ, Battini F, Efthymiou A, Cruz-Paredes C, Nybroe O, Jakobsen I (2018) Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J 12:1296–1307. https://doi.org/10.1038/s41396-018-0059-3
Tanabe T, Funahashi T, Nakao H, Miyoshi SI, Shinoda S, Yamamoto S (2003) Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J Bacteriol 185:6938–6949. https://doi.org/10.1128/JB.185.23.6938-6949.2003
Tani A, Sahin N, Fujitani Y, Kato A, Sato K, Kimbara K (2015) Methylobacterium species promoting rice and barley growth and interaction specificity revealed with whole-cell matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF/MS) analysis. PLoS One 10:e0129509. https://doi.org/10.1371/journal.pone.0129509
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624. https://doi.org/10.1093/nar/gkw569
Team RC (2019) A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. https://www R-project org
Thomashow LS, Weller DM (1988) Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol 170:3499–3508. https://doi.org/10.1128/jb.170.8.3499-3508.1988
Van Loon LC, Bakker PAHM, Pieterse CMJ (1998) Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36:453–483
Varma A, Verma S, Sudha Sahay N, Bütehorn B, Franken P (1999) Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl environ Microbiol 65(6):2741–2744
Venneman J, Audenaert K, Verwaeren J, Baert G, Boeckx P, Moango AM, Dhed’a BD, Vereecke D, Haesaert G (2017) Congolese rhizospheric soils as a rich source of new plant growth-promoting endophytic Piriformospora isolates. Front Microbiol 8:212. https://doi.org/10.3389/fmicb.2017.00212
Verma S, Varma A, Rexer KH, Hassel A, Kost G, Sarbhoy A, Franken P (1998) Piriformospora indica, gen. Et sp. nov., a new root-colonizing fungus. Mycologia 90:896–903. https://doi.org/10.1080/00275514.1998.12026983
Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Huckelhoven R, Neumann C, von Wettstein D, Franken P, Kogel KH (2005) The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. PNAS 102:13386–13391. https://doi.org/10.1073/pnas0504423102
Wang C, Knill E, Glick BR, Défago G (2000) Effect of transferring 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gac a derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can J Microbiol 46:898–907. https://doi.org/10.1139/w00-071
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. https://doi.org/10.1093/nar/gkv437
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exp Bot 52:487–511. https://doi.org/10.1093/jexbot/52suppl_1487
White JF, Kingsley KL, Verma SK, Kowalski KP (2018) Rhizophagy cycle: an oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms 6(3):95. https://doi.org/10.3390/microorganisms6030095
Wickham H, Chang W (2016) W ggplot2: An implementation of the grammar of graphics. https://cranr-projectorg/web/packages/ggplot2/indexhtml
Wickham H, François R, Henry L, Müller K (2019) Dplyr: a grammar of data manipulation. R package version 0.8.3. https://CRANR-projectorg/package=dplyr
Xu XM, Jeffries P, Pautasso M, Jeger MJ (2011) Combined use of biocontrol agents to manage plant diseases in theory and practice. Phytopathology 101:1024–1031. https://doi.org/10.1094/PHYTO-08-10-0216
Zhao Y, Park RD, Muzzarelli RA (2010) Chitin deacetylases: properties and applications. Mar Drugs 8:24–46. https://doi.org/10.3390/md8010024
Zuccaro A, Lahrmann U, Güldener U, Langen G, Pfiffi S, Biedenkopf D, Wong P, Samans B, Grimm C, Basiewicz M, Murat C, Martin F, Kogel KH (2011) Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog 7:e1002290. https://doi.org/10.1371/journalppat1002290
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 676480. We are grateful to Nanna Bygvraa Svenningsen (University of Copenhagen, Denmark) for providing pSEVA237R and assist in the transformation of bacteria, and Philipp Franken, Rosanna C. Hennessy and Maria E. Constantin for providing fungal material. The authors would also like to thank the BestPass consortium for their useful inputs.
Author contribution statement
Alejandro del Barrio-Duque carried out the experiments and wrote the manuscript. Abdul Samad provided expertise in data analysis. Livio Antonielli was responsible for genome assemblage and helped with statistics. Ole Nybroe provided guidance for bacterial transformation. Alejandro del Barrio and Stéphane Compant designed the experiments. All the authors gave also intellectual input and critically revised the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SC is section editor of Plant and Soil. However, this does not interfere with the independent review process.
Additional information
Responsible Editor: Matthew G. Bakker.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
del Barrio-Duque, A., Samad, A., Nybroe, O. et al. Interaction between endophytic Proteobacteria strains and Serendipita indica enhances biocontrol activity against fungal pathogens. Plant Soil 451, 277–305 (2020). https://doi.org/10.1007/s11104-020-04512-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-020-04512-5