Abstract
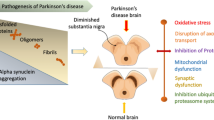
Gaucher’s disease (GD) is a genetic disorder in which glucocerebroside accumulates in cells and specific organs. It is broadly classified into type I, type II and type III. Patients with GD are at high risk of Parkinson’s disease (PD), and the clinical and pathological presentation of GD patients with PD is almost identical to idiopathic PD. Several experimental models like cell culture, animal models, and transgenic mice models were used to understand the molecular mechanism behind GD and PD association; however, such mechanism remains unclear. In this context, based on literature reports, we identified the most common mutations K198T, E326K, T369M, N370S, V394L, D409H, L444P, and R496H, in the Glucosylceramidase (GBA) protein that are known to cause GD1, and represent a risk of developing PD. However, to date, no computational analyses have designed to elucidate the potential functional role of GD mutations with increased risk of PD. The present computational pipeline allows us to understand the structural and functional significance of these GBA mutations with PD. Based on the published data, the most common and severe mutations were E326K, N370S, and L444P, which further selected for our computational analysis. PredictSNP and iStable servers predicted L444P mutant to be the most deleterious and responsible for the protein destabilization, followed by the N370S mutation. Further, we used the structural analysis and molecular dynamics approach to compare the most frequent deleterious mutations (N370S and L444P) with the mild mutation E326K. The structural analysis demonstrated that the location of E326K and N370S in the alpha helix region of the protein whereas the mutant L444P was in the starting region of the beta sheet, which might explain the predicted pathogenicity level and destabilization effect of the L444P mutant. Finally, Molecular Dynamics (MD) at 50 ns showed the highest deviation and fluctuation pattern in the L444P mutant compared to the two mutants E326K and N370S and the native protein. This was consistent with more loss of intramolecular hydrogen bonds and less compaction of the radius of gyration in the L444P mutant. The proposed study is anticipated to serve as a potential platform to understand the mechanism of the association between GD and PD, and might facilitate the process of drug discovery against both GD and PD.






Similar content being viewed by others
References
Abraham MJ, Murtola T, Schulz R et al (2015) GROMACS: high-performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25
Aflaki E, Westbroek W, Sidransky E (2017) The complicated relationship between Gaucher Disease and Parkinsonism: insights from a rare disease. Neuron 93:737–746. https://doi.org/10.1016/j.neuron.2017.01.018
Agrahari AK, Sneha P, George Priya Doss C et al (2017) A profound computational study to prioritize the disease-causing mutations in PRPS1 gene. Metab Brain Dis. https://doi.org/10.1007/s11011-017-0121-2
Agrahari AK, Muskan M, George Priya Doss C et al (2018) Computational insights of K1444N substitution in GAP-related domain of NF1 gene associated with neurofibromatosis type 1 disease: a molecular modeling and dynamics approach. Metab Brain Dis. https://doi.org/10.1007/s11011-018-0251-1
Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R (2004) Mutations in the Glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med 351:1972–1977
Aharon-Peretz J, Badarny S, Rosenbaum H et al (2005) Mutations in the glucocerebrosidase gene and Parkinson disease: phenotype-genotype correlation. Neurology 65:1460–1461
Alcalay RN, Levy OA, Waters CC et al (2015) Glucocerebrosidase activity in Parkinson's disease with and without GBA mutations. Brain 138(Pt9):2648–2658. https://doi.org/10.1093/brain/awv179.
Ali SK, Sneha P, Priyadharshini Christy J et al (2017) Molecular dynamics-based analyses of the structural instability and secondary structure of the fibrinogen gamma chain protein with the D356V mutation. J Biomol Struct Dyn 35:2714–2724
Avnir Y, Prachanronarong KL, Zhang Z et al (2017) Structural determination of the broadly reactive anti-IGHV1-69 anti-idiotypic antibody G6 and its Idiotope. Cell Rep 21:3243–3255
Beavan M, McNeill A, Proukakis C et al (2015) Evolution of prodromal clinical markers of Parkinsondisease in a GBA mutation–positive cohort. JAMA Neurol 72:201. https://doi.org/10.1001/jamaneurol.2014.2950
Bendl J, Stourac J, Salanda O et al (2014) PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol 10:e1003440
Berendsen HJC, Postma JPM, van Gunsteren WF et al (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Beutler E, Gelbart T, Kuhl W (1992) Mutations in Jewish patients with Gaucher disease. Blood 79(7):1662–1666
Brady RO, Kanfer J, Shapiro D (1965) The metabolism of glucocerebrosides. I.Purification and properties of a glucocerebroside-cleaving enzyme from spleen tissue. J Biol Chem 240:39–43
Burrow GA, Barnes S, Grabowski G (2011) Prevalence and management of Gaucher disease. Pediatr Heal Med Ther 2:59. https://doi.org/10.2147/PHMT.S12499
Charrow J, Andersson HC, Kaplan P et al (2000) The Gaucher registry: demographics and disease characteristics of 1698 patients with Gaucher disease. Arch Intern Med 160(18):2835–2843
Chen CW, Lin J, Chu YW (2013) iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinformatics 14:S5. https://doi.org/10.1186/1471-2105-14-S2-S5
Clark LN, Ross BM, Wang Y et al (2007) Mutations in the glucocerebrosidase gene are associated with early-onset Parkinson disease. Neurology 69:1270–1277
Cormand B, Montfort M, Chabas A, Vilageliu L (1997) Genetic fine localization of the beta-glucocerebrosidase (GBA) and prosaposin (PSAP) genes: implications for Gaucher disease. Hum Genet 100:75–79
Dassault Systèmes BIOVIA (2016) Discovery Studio 2016, San Diego: Dassault Systèmes
Doss CGP, Alasmar DR, Bux RI et al (2016) Genetic epidemiology of Glucose-6-phosphate dehydrogenase deficiency in the Arab world. Sci Rep 6:37284
Duran R, Mencacci NE, Angeli AV et al (2013) The glucocerobrosidase E326K variant predisposes to Parkinson’s disease, but does not cause Gaucher’s disease. Mov Disord 28:232–236
Essmann U, Perera L, Berkowitz ML et al (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577
Fernandes HJR, Hartfield EM, Christian HC et al (2016) ER stress and autophagic perturbations lead to elevated extracellular α-synuclein in GBA-N370S Parkinson’s iPSC-derived dopamine neurons. Stem Cell Reports 6:342–356. https://doi.org/10.1016/j.stemcr.2016.01.013
Fishbein I, Kuo Y-M, Giasson BI, Nussbaum RL (2014) Augmentation of phenotype in a transgenic Parkinson mouse heterozygous for a Gaucher mutation. Brain 137:3235–47
Gan-Or Z, Amshalom I, Kilarski LL et al (2015) Differential effects of severe vs mild GBA mutations on Parkinson disease. Neurology 84:880–887
Gaucher PCE (1882) De l'epithelioma primitif de la rate, hypertrophie idiopathique del la rate san leucemie. MD thesis (Paris, France)
Gegg ME, Burke D, Heales SJ et al (2012) Glucocerebrosidase deficiency in substantia nigra of Parkinson disease brains. Ann Neurol 72:455e463
Gegg ME, Sweet L, Wang BH et al (2015) No evidence for substrate accumulation in Parkinson brains with GBA mutations. Mov Disord 30:1085–1089
George Priya Doss C, Zayed H (2017) Comparative computational assessment of the pathogenicity of mutations in the Aspartoacylase enzyme. Metab Brain Dis 32:2105–2118
George Priya Doss C, Chakraborty C, Monford Paul Abishek N et al (2014) Application of evolutionary based in silico methods to predict the impact of single amino acid substitutions in vitelliform macular dystrophy. Adv Protein Chem Struct Biol 94:177–267
Goker-Alpan O, Schiffmann R, LaMarca ME, et al (2004) Parkinsonism among Gaucher disease carriers. J Med Genet 41:937–940
Goker-Alpan O, Lopez G, Vithayathil J et al (2008) The spectrum of parkinsonian manifestations associated with glucocerebrosidase mutations. Arch Neurol 65:1353–1357
Grabowski GA (2008) Phenotype, diagnosis, and treatment of Gaucher’s disease. Lancet 372:1263–1271
Grabowski GA (2012) Gaucher disease and other storage disorders. Hematol Am Soc Hematol Educ Progr 2012:13–18. https://doi.org/10.1182/asheducation-2012.1.13
Grantcharova VP, Riddle DS, Santiago JV et al (1998) Important role of hydrogen bonds in the structurally polarized transition state for folding of the src SH3 domain. Nat Struct Mol Biol 5:714–720
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Hridya H, Amrita A, Mohan S et al (2016) Functionality study of santalin as tyrosinase inhibitor: a potential depigmentation agent. Int J Biol Macromol 86:383–389
Hruska KS, LaMarc ME, Scott CR et al (2008) Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA). Hum Mutat 29:567–583
Keatinge M, Bui H, Menke A et al (2015) Glucocerebrosidase 1 deficient Danio rerio mirror key pathological aspects of human Gaucher disease and provide evidence of early microglial activation preceding alpha-synuclein-independent neuronal cell death. Hum Mol Genet 24:6640–6652
Lesage S, Anheim M, Condroyer C et al (2011) Large-scale screening of the Gaucher’s disease-related glucocerebrosidase gene in Europeans with Parkinson’s disease. Hum Mol Genet 20:202–210
Lieberman RL, D'aquino JA, Ringe D et al (2009) Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability. Biochemistry 48:4816–4827
Linari S, Castaman G (2015) Clinical manifestations and management of Gaucher disease. Clin Cases Miner Bone Metab 12:157–164. https://doi.org/10.11138/ccmbm/2015.12.2.157
Lobanov MI, Bogatyreva NS, Galzitskaia OV (2008) Radius of gyration is indicator of compactness of protein structure. Mol Biol (Mosk) 42:701–706
Magalhaes J, Gegg ME, Migdalska-Richards A et al (2016) Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: relevance to Parkinson’s disease. Hum Mol Genet 25:3432–3445
Mallett V, Ross JP, Alcalay RN et al (2016) GBA p.T369M substitution in Parkinson disease: polymorphismor association? A meta-analysis. Neurol Genet 2:e104. https://doi.org/10.1212/NXG.0000000000000104
Manickam M, Ravanan P, Singh P et al (2014) In silico identification of genetic variants in glucocerebrosidase (GBA) gene involved in Gaucher's disease using multiple software tools. Front Genet 5:148. https://doi.org/10.3389/fgene.2014.00148 eCollection 2014
Mao X, Wang T, Peng R et al (2013) Mutations in GBA and risk of Parkinson’s disease: a meta-analysis based on 25 case-control studies. Neurol Res 35:873–878
Mehta A (2006) Epidemiology and natural history of Gaucher’s disease. Eur J Intern Med 17:S2–S5. https://doi.org/10.1016/j.ejim.2006.07.005
Meshach Paul D, Rajasekaran R (2017) In silico approach to explore the disruption in the molecular mechanism of human hyaluronidase 1 by mutant E268K that directs Natowicz syndrome. Eur Biophys J 46(2):157–169
Meshach Paul D, Rajasekaran R (2018) Exploration of structural and functional variations owing to point mutations in α-NAGA. Interdiscip Sci Comput Life Sci 10:81–92. https://doi.org/10.1007/s12539-016-0173-8
Migdalska-Richards A, Ko WK, Li Q et al (2017) Oral ambroxol increases brain glucocerebrosidase activity in a nonhuman primate. Synapse. https://doi.org/10.1002/syn.21967
Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13:952–962
Mosaeilhy A, Mohamed MM, GPD C et al (2017) Genotype-phenotype correlation in 18 Egyptian patients with glutaric acidemia type I. Metab Brain Dis 32:1417–1426
Moseilhy A, Hassan MM, El Abd HSA et al (2017) Severe neurological manifestations in an Egyptian patient with a novel frameshift mutation in the Glutaryl-CoA dehydrogenase gene. Metab Brain Dis 32:35–40
Nagasundaram N, C GPD, Chakraborty C et al (2016) Mechanism of artemisinin resistance for malaria PfATP6 L263 mutations and discovering potential antimalarials : An integrated computational approach. Nat Publ Gr 1–12
Naine SJ, Devi CS, Mohanasrinivasan V et al (2015) Binding and molecular dynamic studies of sesquiterpenes (2R-acetoxymethyl-1,3,3-trimethyl-4t-(3-methyl-2-buten-1-yl)-1t-cyclohexanol) derived from marine Streptomyces sp. VITJS8 as potential anticancer agent. Appl Microbiol Biotechnol 1–14
Offman MN, Krol M, Silman I et al (2010) Molecular basis of reduced glucosylceramidase activity in the most common Gaucher disease mutant, N370S. J Biol Chem 285:42105–42114
O’Regan G, deSouza R-M, Balestrino R, Schapira AH (2017) Glucocerebrosidase mutations in Parkinson Disease. J Parkinsons Dis 7:411–422. https://doi.org/10.3233/JPD-171092
Ou L, Przybilla MJ, Whitley CB (2017) Phenotype prediction for mucopolysaccharidosis type I by in silico analysis. Orphanet J Rare Dis 12:125. https://doi.org/10.1186/s13023-017-0678-1
P S, D KT, Tanwar H et al (2017) Structural analysis of G1691S variant in the human Filamin B gene responsible for Larsen syndrome: a comparative computational approach. J Cell Biochem 118:1900–1910
Pace CN, Fu H, Lee Fryar K et al (2014) Contribution of hydrogen bonds to protein stability. Protein Sci 23:652–661
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190
Petrova SS, Solov’ev AD (1997) The origin of the method of steepest descent. Hist Math 24:361–375
Rodriguez-Porcel F, Espay AJ, Carecchio M (2017) Parkinson disease in Gaucher disease. J Clin Mov Disord 4:7. https://doi.org/10.1186/s40734-017-0054-2
Sahu VK, Khan AKR, Singh RK et al (2008) Hydrophobic, polar and hydrogen bonding based drug-receptor interaction of tetrahydroimidazobenzodiazepinones. Am J Immunol 4:33–42
Sanchez-Martinez A, Beavan M, Gegg ME et al (2016) Parkinson’s disease-linked GBA mutation effects reversed by molecular chaperones in human cell and fly models. Sci Rep 6:31380
Sanders A, Hemmelgarn H, Melrose HL et al (2013) Transgenic mice expressing human glucocerebrosidase variants: utility for the study of Gaucher disease. Blood Cells Mol Dis 51:109–115
Sardi SP, Cheng SH, Shihabuddin LS (2015) Gaucher-related synucleinopathies: the examination of sporadic neurodegeneration from a rare (disease) angle. Prog Neurobiol 125:47–62
Schmid N, Eichenberger AP, Choutko A et al (2011) Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 40:843–856
Schneider JP, Kelly JW (1995) Templates that Induce .Alpha.-helical, .Beta.-sheet, and loop conformations. Chem Rev 95:2169–2187
Sidransky E (2012) Gaucher disease: insights from a rare Mendelian disorder. Discov Med 14:273–281
Sidransky E, Nalls MA, Aasly JO et al (2009) Multicenter analysis of Glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med 361:1651–1661
Sievers F, Higgins DG (2014) Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079:105–116
Smith L, Mullin S, Schapira AHV (2017) Insights into the structural biology of Gaucher disease. Exp Neurol 298(Pt B):180–190
Sneha P, Priya Doss CG (2016) Molecular dynamics: new frontier in personalized medicine. Adv Protein Chem Struct Biol 102:181–224
Sneha P, Thirumal KD, George PDC et al (2017a) Determining the role of missense mutations in the POU domain of HNF1A that reduce the DNA-binding affinity: a computational approach. PLoS One 12:1–24
Sneha P, Thirumal Kumar D, Saini S et al (2017b) Analyzing the effect of V66M mutation in BDNF in causing mood disorders: a computational approach. Adv Protein Chem Struct Biol 108:85–103
Sneha P, Ebrahimi EA, Ghazala SA et al (2018) Structural analysis of missense mutations in galactokinase 1 (GALK1) leading to galactosemia type-2. J Cell Biochem. https://doi.org/10.1002/jcb.27097
Sujitha SP, Kumar DT, Doss CGP et al (2016) DNA repair gene (XRCC1) polymorphism (Arg399Gln) associated with schizophrenia in south Indian population: a genotypic and molecular dynamics study. PLoS One 11:e0147348
Tanwar H, George Priya Doss C (2018) An integrated computational framework to assess the mutational landscape of α-L-Iduronidase IDUA gene. J Cell Biochem 119(1):555–565. https://doi.org/10.1002/jcb.26214
Tanwar H, Sneha P, Thirumal Kumar D et al (2017) A computational approach to identify the biophysical and structural aspects of methylenetetrahydrofolate reductase (MTHFR) mutations (A222V, E429A, and R594Q) leading to Schizophrenia. Adv Protein Chem Struct Biol 108:105–125
Tayebi N, Walker J, Stubblefield B, et al (2003) Gaucher disease with parkinsonian manifestations: does glucocerebrosidase deficiency contribute to a vulnerability to parkinsonism? Mol Genet Metab 79:104–9
Thirumal Kumar D, George Priya Doss C, Sneha P et al (2016) Influence of V54M mutation in giant muscle protein titin: a computational screening and molecular dynamics approach. J Biomol Struct Dyn 1102:1–12
Thirumal Kumar D, George Priya Doss C, Sneha P et al (2017a) Influence of V54M mutation in giant muscle protein titin: a computational screening and molecular dynamics approach. J Biomol Struct Dyn 35:917–928
Thirumal Kumar D, Lavanya P, George Priya Doss C et al (2017b) A molecular docking and dynamics approach to screen potent inhibitors against Fosfomycin resistant enzyme in clinical Klebsiella pneumoniae. J Cell Biochem. https://doi.org/10.1002/jcb.26064
Thirumal Kumar D, Jerushah Emerald L, George Priya Doss C et al (2018) Computational approach to unravel the impact of missense mutations of proteins (D2HGDH and IDH2) causing D-2-hydroxyglutaric aciduria 2. Metab Brain Dis. https://doi.org/10.1007/s11011-018-0278-3
Ullah I, Nasir A, Mehmood S et al (2017) Identification and in silico analysis of GALNS mutations causing Morquio a syndrome in eight consanguineous families. Turk J Biol 41:458–468
Wei RR, Hughes H, Boucher S et al (2011) X-ray and biochemical analysis of N370S mutant human acid β-glucosidase. J Biol Chem 286:299–308
Xu YH, Quinn B, Witte D et al (2003) Viable mouse models of acid beta-glucosidase deficiency: the defect in Gaucher disease. Am J Pathol 163:2093–2101
Yagawa K, Yamano K, Oguro T et al (2010) Structural basis for unfolding pathway-dependent stability of proteins: Vectorial unfolding versus global unfolding. Protein Sci 19:693–702
Yang S-Y, Beavan M, Chau K-Y et al (2017) A human neural crest stem cell-derived dopaminergic neuronal model recapitulates biochemical abnormalities in GBA1 mutation carriers. Stem Cell Reports 8:728–742. https://doi.org/10.1016/j.stemcr.2017.01.011
Yun S, Guy HR (2011) Stability tests on known and misfolded structures with discrete and all atom molecular dynamics simulations. J Mol Graph Model 29:663–675. https://doi.org/10.1016/j.jmgm.2010.12.002
Yun SP, Kim D, Kim S et al (2018) α-Synuclein accumulation and GBA deficiency due to L444P GBA mutation contributes to MPTP-induced parkinsonism. Mol Neurodegener 13(1):1. https://doi.org/10.1186/s13024-017-0233-5
Zaki OK, El Abd HS, Mohamed SA et al (2016) Novel mutation in an Egyptian patient with infantile Canavan disease. Metab Brain Dis 31:573–577
Zaki OK, Krishnamoorthy N, El Abd HS et al (2017a) Two patients with Canavan disease and structural modeling of a novel mutation. Metab Brain Dis 32:171–177
Zaki OK, Priya Doss CG, Ali SA et al (2017b) Genotype-phenotype correlation in patients with isovaleric acidaemia: comparative structural modelling and computational analysis of novel variants. Hum Mol Genet 12:e0174953
Zuckerman S, Lahad A, Shmueli A et al (2007) Carrier screening for Gaucher disease: lessons for low-penetrance, treatable diseases. JAMA 298(11):1281–1290
Acknowledgments
The authors acknowledge the management of Vellore Institute of Technology for the seed money and (BRAF) @ CDAC for providing the facilities required to perform this work.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure statement
No potential conflict of interest was reported by the authors.
Electronic supplementary material
ESM 1
(DOCX 233 kb)
Rights and permissions
About this article
Cite this article
Thirumal Kumar, D., Eldous, H.G., Mahgoub, Z.A. et al. Computational modelling approaches as a potential platform to understand the molecular genetics association between Parkinson’s and Gaucher diseases. Metab Brain Dis 33, 1835–1847 (2018). https://doi.org/10.1007/s11011-018-0286-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-018-0286-3