Abstract
Campylobacter jejuni is a spiral-shaped Gram-negative and microaerophilic bacteria that causes bacterial diarrhea worldwide. Due to its complicated antigenic nature, lack of proper animal models, and a poor understanding of its pathogenicity there is no vaccine against C. jejuni. The most effective way of reducing the number of Campylobacter infections is by developing protective vaccines for humans. Due to the increasing resistance against antibiotics of bacterial strains epitopes-based vaccine could be the best approach to trigger an effective immune response. Immunoinformatics methods can be an alternative approach for developing epitope-based vaccine which is able to elucidate humoral and cell-mediated immune response inside the host body. Reverse vaccinology approach analyses the entire protein sequences of the pathogen using bioinformatics tools to select target proteins for their high-throughput expression and validation. Here in this research study, three epitopes MSNVYAYRF, LSDDINLNI and ATSTSTITL have been identified as the most potential epitopes binding with HLA-B*58:01, HLA-A*01:01 and HLA-B*07:02 MHC class I allele, respectively. With the help of Autodock vina, these three epitopes have shown the lowest binding energies and form the most complex docked structure covering the maximum number of populations worldwide. Thus, we suggest these peptides may have a good potential if further presented for experimental analysis and can be proved to be helpful against C. jejuni infections causing diarrhea.
Similar content being viewed by others
Introduction
Campylobacter jejuni is a Gram-negative spiral-shaped bacteria which is one of the leading causes of foodborne bacterial diseases worldwide. The isolation of C. jejuni was accomplished from human feces in 1968 (Dekeyser et al. 1972). Since its discovery, C. jejuni remains the most frequent cause of infectious diarrhea affecting over 450 million people every year throughout the world, attributing a large economic burden (Friedman et al. 2000). It is most commonly found in animal feces and therefore poultry is the major reservoir (Young et al. 2007). Diarrhea is the third leading cause of childhood mortality in India and it is responsible for 13% of all death per year in children under 5 year of age killing in an estimated 300,000 children per year in India. Campylobacter infection affects more than 1.3 million people every year (CDC 2017). According to WHO report, diarrheal diseases are the most common infection resulting from unhygienic food causing 550 million people falling ill yearly out of which 220 million children below the age of 5 years and it is one of the key global causes of diarrheal diseases (WHO 2018). C. jejuni infects the organism by penetrating the gastrointestinal mucus, then adhering to the gut enterocytes and induced diarrhea by toxin release mainly enterotoxin and cytotoxin that correlate with the severity of enteritis (Gallardo et al. 1998; Wallis 1994). The limited data on the treatment of C. jejuni infection suggested that ciprofloxacin, erythromycin, and fluoroquinolones may shorten the duration of symptoms. However, treatment failure associated with the emergence of the quinolone-resistant strain of C. jejuni has been documented (Van et al. 2003). For the permanent solution with the infection of C. jejuni, there is need of a powerful vaccine that could provide immunity against it but till now no vaccine is available against campylobacter.
Reverse vaccinology is the best approach in terms of medical sciences that involves the use of genomic information with the use of computer for the preparation of vaccines without culturing microorganism (Kanampalliwar et al. 2013; Tang et al. 2012). This method helps in the development of vaccines that were previously difficult or impossible to make and therefore can lead to the discovery of the unique epitope-based vaccine that will improve existing vaccine technology (Sette and Rappuoli 2010). It allows the selection of antigenic regions from the pathogenic genomes and the most antigenic regions could be developed as potential vaccine candidates to trigger protective immune responses (Ada et al. 2018). Production of peptide or epitopes based vaccines are easy in comparison to the conventional vaccine and it is also more specific, cost-effective, less time consuming and safe (Kumar et al. 2015). An epitope-based vaccine recently drawn much attention in treating infectious diseases (Yang and Yu 2009). Immunization based on epitope-based vaccines is more powerful in stimulation of the cellular and humoral immune response (Bijker et al. 2007). These types of vaccines consist of highly immunogenic T and B cells epitopes, which provoke cytotoxic T cells (CTL), TH or B to specific epitopes (Baloria et al. 2012; Akhoon et al. 2011). B and TH cells play an important role in the induction of a protective immune response in many bacterial infections; thus, determination of peptides that induce T and B cells response is the crucial requirement for the design of effective epitope-based vaccines (Gupta et al. 2010, 2012). Due to the heterogeneous virulence nature of C. jejuni infection, there is a lack of proper animal models that can replicate the gastroenteritis caused by C. jejuni in humans. C. jejuni vaccine candidates that have been or are in clinical development include killed the whole cell, protein subunit, and capsule-conjugate products (Man 2011). A recombinant protein vaccine by the name ACE 393 has been tested in Phase II human challenge study but its efficacy failed to demonstrate protection (Riddle and Guerry 2016). To validate the analysis, two standard antigens were taken as control (CadF and FlpA) which helps in the colonization of the gut that is promoted by flagellum-mediated motility and binding to host tissue such as fibronectin (Flanagan et al. 2009; Jin et al. 2001).
In the present study, screening of the whole genome in search of probable antigens was performed by using the RV approach. Immuno-informatics or computational biology has added an unavoidable contribution to design epitope-based vaccines (Gupta et al. 2011). In this context, identification of potential epitopes from an antigen protein by in silico methods can be considered in such vaccines reducing the lengthy process for discovery of appropriate epitopes (Srivastava et al. 2011). Instead of selecting individual proteins, the RV approach analyses the entire protein sequences of the pathogen using bioinformatics tools to select target proteins for their high-throughput expression and validation in animal models. The wide application of computational method provides an easy way to identify the epitopes of highly antigenic protein. Vaxign, a server that predicts the novel antigen which can be used as potential candidates for designing vaccine (Yongqun and Mobley 2010). Predicted epitopes will be further examined for different physiochemical and immunogenic properties as well as three-dimensional structural modeling and the docking studies will be performed to investigate the strong binding interaction with MHC I alleles. Nowadays, many online servers are available for predicting B and T cell epitopes. In this respect, the Immune Epitope Database (IEDB) server website (Vita et al. 2010) provides tools to predict both B and T cells epitopes. Other online servers such as ProPred (Singh and Raghava 2001), MHCpred (Guan et al. 2003) and SVMHC (Donnes and Elofsson 2002) have different tools for finding T cell epitopes. In order to examine the binding capability of these predicted epitopes with their corresponding alleles and population coverage analysis were performed.
Methodology
The C. jejuni strain NCTC 11,168 has a circular chromosome of 1,641,481 base pairs (30.6% G+C) that encodes 1680 proteins (Parkhill et al. 2000). The most striking feature of this strain was the presence of hypervariable sequences and lack of repetitive DNA sequences within this genome. The steps involved in the screening of the complete genome have been portrayed in Fig. 1.
Retrieval of Proteome Data and Identification of Antigens
The complete proteome of diarrheagenic campylobacter jejuni strain NCTC 11,168 was isolated from UniProt database having proteome id UP000000799 (http://www.uniprot.org/proteomes/) showing the total number of 1680 protein coding sequences with their complete protein information. Adhesin Probability > = 0.51 of the filtered antigens are screened from the complete proteome of Campylobacter jejuni through Vaxign Server (Xiang and He 2009) (http://www.violinet.org/vaxign/index.php). Vaxign is a web-based software that works on the principle of reverse vaccinology and predicts the vaccine targets by computing the complete genome using default parameters. This server uses the default cutoff value of 0.51 to predict adhesion probability of the vaccine candidate. The adhering capability of the pathogenic strain is an important characteristic in causing infection and therefore considered to be the major target in the development of vaccine.
Prediction of Antigenic and Allergic Properties
Selected proteins were further subjected to VaxiJen tool for the characterization of the antigenic nature of protein (Doytchinova et al. 2007). VaxiJen is the first server to predict the protective antigens mainly based on their physicochemical properties of proteins irrespective of the sequence alignment. In order to achieve high accuracy of prediction, only those proteins that show above 0.51 cut off scores were selected and therefore 49 proteins were calculated for analysis. These antigenic proteins sequences were also tested for their allergenicity prediction by using Allergen FP v.1.0 tool available at http://www.ddg-pharmfac.net/AllergenFP/index.html. It is a novel fingerprint based approach that differentiates between the allergens and non-allergens (Dimitrov et al. 2014).
Prediction of T Cell Epitopes
The binding capability of the T cell epitopes with HLA class I alleles were checked by using NetCTL 1.2 tool (Larsen et al. 2007) and Net MHC pan 4.0 (Vanessa et al. 2017). NetCTL 1.2 server predicts CTL epitopes in any given protein sequences, proteasomal C terminal cleavage and TAP transport efficiency. This server allows for the predictions of CTL epitopes restricted to 12 MHC class I supertype. It uses the different algorithm for prediction of MHC class I binding and proteasomal cleavage by using artificial neural networks and TAP transport efficiency prediction through weight matrix method. To confirm the predicted peptides analyze through NetCTL 1.2 server it was further validated by using Net MHC pan 4.0. This server predicts binding of peptides to an MHC molecule of the antigenic sequence using artificial neural networks (ANNs).
To confirm the most probable epitopes with the higher confidence level, the final peptides were analyzed again through VaxiJen v2.0 web server and peptides that have shown high scores were selected for the analysis. High scorer peptides showing score > = 1.1 were shortlisted for the toxicity prediction. On the basis of these scores, best vaccine candidates were selected for further analysis.
Characterization of Target Proteins as a Vaccine Candidate
SPAAN is a non-homology based software program that is based on the properties of the sequence presented. It identifies top-scoring novel adhesins with the high level of confidence, therefore, the adhesion property of the antigens was checked by SPAAN (Sachdeva et al. 2005). Localization of these proteins was predicted by PSORT b 3.0 (Yu et al. 2010). PSORTb has remained the most precise bacterial protein, subcellular localization (SCL) predictor which analyzed the subcellular localization by using multiple analytical modules. Prediction of the subcellular localization of proteins through computational approach is a valuable tool for genome analysis and annotation and it can provide information regarding its function in an organism. Transmembrane regions were also checked by TMHMM method (Krogh et al. 2001). TMHMM is a membrane protein topology prediction method and works on the principle of hidden Markov model. It predicted the number of transmembrane helices within the protein sequences and discriminate between soluble and membrane proteins with the high level of accuracy.
Prediction of Physiochemical Properties of Proteins
Identification of physical and chemical properties have been performed by ProtParam tool (Gasteiger et al. 2005). These parameters include molecular weight, theoretical pI, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity. ABTMpro is a server that characterizes whether a given protein sequence is a transmembrane protein or not. It further identifies the probabilities of the transmembrane protein predicted by ABTMpro being an alpha-helical transmembrane protein or a Beta Barrel transmembrane protein. It consists of a Support Vector Machine, which utilizes features such as amino acid composition and properties, reduced alphabet composition, predicted secondary structure, evolutionary information etc. ABTM pro is available at http://scratch.proteomics.ics.uci.edu/.
IEDB Percentile Score Calculation of Predicted Epitopes
Binding affinities of the peptides were calculated by IEDB tool by applying Stabilized Matrix Method against the MHC restricted HLA alleles (Peters and Sette 2005). Toxicity of all the selected peptides were examined by Toxin Pred tool (Gupta et al. 2013). Toxin Pred is an in silico tool that predicts and design the toxic as well as non-toxic peptides. Here, in our study, we require non-toxic peptides, therefore, computational methods are useful in predicting the toxicity of peptides but also facilitate the designing of better useful peptides with lower toxicity level (Khan et al. 2017). This tool not only discriminates against toxic peptides from non-toxic peptides but also save time and money.
Molecular Modeling of HLA Alleles and Epitopes
The three-dimensional structure of the epitopes was designed by using PEPstrMOD (Kaur et al. 2007), a server that predicts the 3D structures of the small peptide as well as peptides having non-natural residues, terminal modifications, cyclization, and post-translational modifications, etc. The sequence of HLA alleles was downloaded from IPD-IMGT/HLA Database (Robinson et al. 2015). The 3-D structures of these HLA alleles were modeled by Modeller 9.18 (Sali et al. 1995). MODELLER is used for homology or comparative modeling of the three-dimensional structures of the proteins and provides an alignment of a protein sequence to be modeled with their known structural template. It automatically calculates five models and the best of all model was selected on the basis of their lowest DOPE score. The most acceptable model was further validated by RAMPAGE and ProSA analysis (Wiederstein and Sippl 2007). This tool is widely used to check 3D models of protein structures for potential errors.
Molecular Docking Analysis of the Peptide and HLA Allele Complex
After structural modeling, docking of the selected peptides with their favored HLA alleles was performed by using AutoDock Vina (Trott and Olson 2010). The tool generates 9 docked conformations that were ranked on the basis of binding energy, torsion energy, geometry and electrostatic energy. The best output was finalized on the basis of highest binding energy score. AutoDock Vina did considerably better in terms of speed and accuracy. Preparation of ligand and protein was done by using Autodock 4.2 of MGL tool 1.5.4. The chimera tool was used for the visualization of the 3-D structure with their Binding position, H-bonding between the HLA alleles and peptides.
Worldwide Population Coverage Analysis
The frequency of distribution of human HLA alleles binding to MHC molecules was predicted by MHC Pred (Guan et al. 2003) and their conservancy analysis among the predicted epitopes was carried out by IEDB population coverage tool (Bui et al. 2006). All the parameters were set to default values that have a dataset containing the total number of 3245 alleles for 16 geographical areas, 21 ethnicities, and 115 countries. The predicted antigenic epitopes with their corresponding HLA alleles were submitted to this database and it determines the percentage of the number of population interacting to a given set of epitopes.
Results and Discussion
Identification of Protective Antigens and Antigenicity Prediction
In the present study, the screening of complete proteome of C. jejuni in order to identify protective antigens with higher affinity was done by Vaxign server. Identification of a set of 82 proteins was shortlisted on the basis of adhesion value above 0.51 which indicates the adhesion properties of the filtered proteins. Subsequently, the next step is the filtering of predicted proteins via VaxiJen to select only those sequences having the high probability of antigenic epitopes. Out of 82 proteins, 49 proteins were predicted as potential antigens and furthermore can be characterized by their allergic properties with AllergenFP tool to exclude the allergens proteins. The non-allergens with high VaxiJen scores above 0.8 were selected for the further analysis- Q0PB90, Q0PB82, Q0PA31, Q0P914, Q0P8F1 (Table 1).
Characterization of Target Antigens as Potential Vaccine Candidates
To characterize predicted antigens as good vaccine candidates, we have applied various computational methods to distinguish their immunological properties (Khan et al. 2018). The SPAAN program (Sachdeva et al. 2005) is applied to calculate the adhesion property of the target antigens, all the 5 proteins were predicted as adhesions. Outer membrane surface proteins were considered to be a good choice for vaccine development and so the prediction of localization of these proteins was done by PSORT b 3.0. Usually, proteins having more than two transmembrane region are not considered a good vaccine candidate due to the difficulty in cloning expressing and purifying. Here we had used the TMHMM method which clearly depicted that all the 5 selected proteins were having less than 1 transmembrane region. Since it is difficult to clone, express and purify protein antigens with 1 or more TM region and is a major obstacle in the potential vaccine candidates selection. Furthermore, ABTM pro was applied to predict alpha helix and beta barrel transmembrane proteins with the overall accuracy of 97% (Singh et al. 2017) (Table 2).
Prediction and Analysis of MHC Restricted T Cell Epitopes
Epitopes that are capable of inducing T-cell immune response are considered to be a good vaccine candidate. In the recent years, all the efforts have been devoted to the identification of MHC restricted T cell epitopes (Singh et al. 2010). For the identification of these epitopes, amino acid sequences of all the predicted proteins were subjected to Net CTL 1.2 for T- cell epitope prediction keeping the combined epitope prediction score of 0.75 (Tables 3 and 4). Thus, 5 predicted antigens sequences along with two control proteins were screened for their promiscuous HLA class I-restricted supertype binding epitopes using the NetCTL algorithm. The control antigen proteins were included in the study to compare the epitope density/immunogenicity level of the predicted antigens. Cytotoxic T cells are considered to be important in the immune system’s response to disease as they have the capability to recognize defective cells by binding to peptides presented on the cell surface by MHC (Major Histocompatibility Complex) class I molecules (Huber et al. 2014). To validate the peptides predicted through the NetCTL algorithm another latest more precise server Net MHC pan 4.0 were applied and results were compared by comparing the peptides scores. The threshold for the strong binders was kept at its default value 0.5 and therefore peptides showing below this values were considered to be the weak binder (Tables 5 and 6). In addition, the selected epitopes were further subjected to VaxiJen server in order to predict their antigenicity to elicit an effective immune response. As the epitopes displaying the top scores > = 1.1 were selected and low score binders were eliminated as shown in Table 7.
We have presented all the selected 7 epitopes to MHC Pred and IEDB to predict the binding energies to their corresponding HLA alleles. Peptides binding to MHC class I alleles (IC50) ≤ 500 nM were selected for the further analysis. The solubility of all the selected epitopes was predicted by the ProtParam tool and molecular weight, as well as other properties, were mentioned in Table 8.
The result from toxicity prediction clearly shows that all the selected antigenic epitopes are non-toxic in nature (Table 9).
Modeling and Refinement of the Identified Peptides and MHC Alleles
The three-dimensional structural information of proteins plays a significant role in providing the insight information on their molecular functions and identification of their binding sites. Mapping of the 3-D structure of the MHC I HLA alleles—HLA-A*01:01, HLA-A*24:03, HLA-B*07:02, HLA B*39:01, and HLA-*58:01 with respect to its highest scoring epitopes were generated by Modeller 9.18 which used the template base model that was retrieved from Protein Data Bank. In order to refine the quality of structures generated through MODELLER, these were further subjected to RAMPAGE tool. The analysis indicated that all the modeled structure’s residues were > 90% in their respective favored region, and therefore approved the quality of the predicted models. Furthermore, the three selected protein models were validated through ProSA analysis (Fig. 2). Now it came to light that only segments of antigenic protein or epitopes were enough to induce the desired immune response in comparison to the whole protein (Huber et al. 2014).
ProSA analysis all selected protein chains in PDB determined by X-ray crystallography (light blue) or NMR spectroscopy (dark blue) with respect to their length. a HLA-A*01:01 allele representing − 7.27 Z score b HLA-B*07:02 allele representing − 7.17 Z score c HLA-B*58:01 allele representing − 7.92 Z score
Binding Energy Calculation of the HLA Allele-Epitope Complex
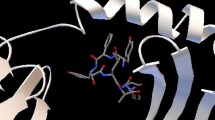
Using Autodock vina, the docking of the predicted set of epitopes and HLA allele was performed. Different ten conformations were generated by Autodock vina showing the binding interaction and the best conformation was selected on the basis of binding energy score. The lower binding energy is stronger the bonding interaction between HLA allele and epitope. The protein and the ligand preparation was done by using Autodock 4.2 tool and PDB files were converted into the PDBQT files required for docking through Autodock vina. The interactions between MSNVYAYRF binding with HLA-B*58:01, LSDDINLNI with HLA-A*01:01 and ATSTSTITL with HLA-B*07:02 are shown in Table 10. The binding energies are − 9.2, − 7.6 and − 7.4 kcal/mol, respectively. Along with these epitopes, two best peptides from the control antigens were also docked to compare the docking result, ITPTLNYNY(+) shown − 7.7 score and KANASISIK(+) shown − 7.5 score respectively. Three epitopes with the best energies were selected and were analyzed by Chimera 1.2 tool and docked complex was generated and visualized through Chimera as shown in Figs. 3, 4 and 5.
Worldwide Population Coverage Analysis
Population coverage analysis was performed for selected epitopes along with their corresponding MHC-I alleles predicted through IEDB tool. The three predicted epitopes MSNVYAYRF, LSDDINLNI and ATSTSTITL have shown the immune response elicitation of the 56.35%, 42.41%, and 53.11% total world population. Maximum population coverage was for observed for epitope MSNVYAYRF to be 70.91% (Fig. 6), epitope LSDDINLNI to be 56.39% (Fig. 7) and epitope ATSTSTITL to be 66.85% (Fig. 8) were found in the population of Northeast Asia. These epitopes have shown no population coverage in the region of South Africa. The results from population coverage analysis analyzed through IEDB tool clearly indicate the distribution of epitopes in the different region.
Conclusion
The computational approach can be the helpful method in predicting the best epitopes on the basis of their antigenic nature, non-toxigenic score, interacting with the maximum number of HLA alleles and with higher population coverage in vaccine development. Considering all the above parameter, three epitopes MSNVYAYRF, LSDDINLNI and ATSTSTITL predicted to have the most considerable binding with HLA-B*58:01, HLA-A*01:01 and HLA-B*07:02 MHC class I allele and lowest binding energy values providing stability to the peptide and MHC complex. Therefore, these peptide candidates can be further suggested for experimental laboratory analysis for the development of an effective vaccine against diarrhea. Screenings through experimental methods are time-consuming and laborious and therefore can be replaced by reverse vaccinology based immunoinformatics approach. Therefore, the current study used in silico methods to reduce the time-consuming laboratory experiments and to avoid a hit and trial method. Thus, our findings can be helpful in developing vaccines against C. jejuni infection after experimentally tested in the laboratory.
References
Ada K, Candy C, Abu B, Abdul M, Chiuan H, Boon HL, Chiuan YL (2018) Current progress of immunoinformatics approach harnessed for cellular- and antibody-dependent vaccine design. Pathogens Global Health 112:3
Akhoon BA, Slathia PS, Sharma P, Gupta SK, Verma V (2011) In silico identification of novel protective VSG antigens expressed by Trypanosoma brucei and an effort for designing a highly immunogenic DNA vaccine using IL-12 as adjuvant. Microb Pathog 51(1–2):77–87. https://doi.org/10.1016/j.micpath.2011.01.011
Baloria U, Akhoon BA, Gupta SK, Sharma S, Verma V (2012) In silico proteomic characterization of human epidermal growth factor receptor 2 (HER-2) for the mapping of high affinity antigenic determinants against breast cancer. Amino Acids 42(4):1349–1360. https://doi.org/10.1007/s00726-00010-00830-x
Bijker Melief CJ, Offringa R, van der Burg SH (2007) Design and development of synthetic peptide vaccines: past, present and future. Expert Rev Vac 6(4):591–603. https://doi.org/10.1586/14760584.6.4.591
Bui H, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A (2006) Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform 7:153
Dekeyser P, Gossuin-Detrain M, Butzler JP, Sternon J (1972) Acute enteritis due to related vibrio: first positive stool cultures. J Infect Dis 125:390–392
Dimitrov I, Naneva L, Doytchinova I, Bangov I (2014) AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics 30:846–851
Donnes P, Elofsson A (2002) Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinform 3:25
Doytchinova IA, Darren R, Flower (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8:4
Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME (2009) Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect Immun 77(6):2399–2407
Friedman CR, Neimann J, Wegener HC, Tauxe RV (2000) Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Campylobacter. ASM International, Washington, pp 121–138
Gallardo F, Gascón J, Ruiz J, Corachan M, Jimenez de Anta M, Vila J (1998) Campylobacter jejuni as a cause of traveler’s diarrhea: clinical features and antimicrobial susceptibility. J Travel Med 5(1):23–26
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, pp 571–607
Guan P, Doytchinova IA, Zygouri C, Flower DR (2003) MHCPred: a server for quantitative prediction of peptide—MHC binding. Nucleic Acids Res 31:3621–3624
Gupta SK, Smita S, Sarangi AN, Srivastava M, Akhoon BA, Rahman Q, Gupta SK (2010) In silico CD4? T-cell epitope prediction and HLA distribution analysis for the potential proteins of Neisseria meningitidis serogroup B: a clue for vaccine development. Vaccine 28(43):7092–7097. https://doi.org/10.1016/j.vaccine.2010.08.005
Gupta SK, Srivastava M, Akhoon BA, Smita S, Schmitz U, Wolkenhauer O, Vera J, Gupta SK (2011) Identification of immunogenic consensus T-cell epitopes in globally distributed influenza-A H1N1 neuraminidase. Infect Genet Evol 11(2):308–319. https://doi.org/10.1016/j.meegid.2010.10.013
Gupta SK, Srivastava M, Akhoon BA, Gupta SK, Grabe N (2012) In silico accelerated identification of structurally conserved CD8? and CD4? T-cell epitopes in high-risk HPV types. Infect Genet Evol 12(7):1513–1518. https://doi.org/10.1016/j.meegid.2012.02.022
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8:e73957. https://doi.org/10.1371/journal.pone.0073957
Huber SR, van Beek J, de Jonge J, Luytjes W, van Baarle D (2014) T Cell responses to viral infections—opportunities for peptide vaccination. Front Immunol 5:171
Jin S, Joe A, Lynett J, Hani EK, Sherman P, Chan VL (2001) JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol Microbiol 39(5):1225–1236
Kanampalliwar AM, Soni R, Girdhar A, Tiwari A (2013) Reverse vaccinology: basics and applications. J Vaccines Vaccin. https://doi.org/10.4172/2157-7560.1000194
Kaur H, Garg A, Raghava GPS (2007) PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept Lett 14:626–630
Khan F, Srivastava V, Kumar A (2017) Epitope based peptide prediction from proteome of enterotoxigenic E. coli. Int J Peptide Res Ther https://doi.org/10.1007/s10989-017-9617-1
Khan F, Srivastava V, Kumar A (2018) Computational identification and characterization of potential T-cell epitope for the utility of vaccine design against enterotoxigenic Escherichia coli https://doi.org/10.1007/s10989-018-9671-3
Krogh A, Larsson B, Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Kumar A, Hays M, Lim F, Foster LJ, Zhou M, Zhu G, Miesner T (2015) Protective enterotoxigenic Escherichia coli antigens in a murine intranasal challenge model. PLOS. https://doi.org/10.1371/journal.pntd.0003924
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform 8:424
Man SM (2011) The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol 8(12):669–685
Parkhill J, Wren BW, Mungall KL, Ketley JM, Churcher CM, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rutherford KM, Barrell BG (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000:665–668
Peters B, Sette A (2005) Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform 31 6:132
Riddle S, Guerry P (2016) Status of vaccine research and development for Campylobacter jejuni. Vaccine 34(26):2903–2906
Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE (2015) The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43:D423–D431
Sachdeva G, Kumar K, Jain P, Ramachandran S (2005) SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics 21:483–491
Sali A, Potterton L, Feng Y, Herman V, Martin K (1995) Evaluation of comparative protein modeling by Modeller. Protein 23:318–326
Sette A, Rappuoli R (2010) Reverse vaccinology: developing vaccines in the era of genomics. Immunity 33(4):530–541
Singh H, Raghava GPS (2001) ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236–1237
Singh SP, Deeksha S, Bhartendu NM (2017) Genome-wide identification of novel vaccine candidates for Plasmodium falciparum malaria using integrative bioinformatics approaches. 3 Biotech 7:318
Singh SP, Khan F, Mishra BN (2010) Computational characterization of Plasmodium falciparum proteomic data for screening of potential vaccine candidates. Hum Immunol 71(2):136–143
Srivastava M, Gupta SK, Abhilash PC, Singh N (2011) Structure prediction and binding sites analysis of curcin protein of Jatropha curcas using computational approaches. J Mol Model 18(7):2971–2979. https://doi.org/10.1007/s00894-011-1320-0
Tang H, Liu XS, Fang YZ, Pan L, Zhang ZW, Zhou P, Lv JL, Jiang ST, Hu WF, Zhang P, Wang YL, Zhang YG (2012) The epitopes of foot and mouth disease. Asian J Anim Vet Adv 7:1261–1265
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Vanessa J, Paul S, Andreatta M, Marcatili P, Peters B, Morten N (2017) NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol https://doi.org/10.4049/jimmunol.1700893
Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B (2010) The immune epitope database 2.0. Nucleic Acids Res 38:D854–D862
Wallis MR (1994) The pathogenesis of Campylobacter jejuni Br J Biomed Sci 51(1):57–64
Wiederstein and Sippl (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410
Xiang Z, He Y (2009) Vaxign: a web-based vaccine target design program for reverse vaccinology. Procedia Vaccinol 1(1):23–29
Yang X, Yu X (2009) An introduction to epitope prediction methods and software. Rev Med Virol 19(2):77–96. https://doi.org/10.1002/rmv.602
Yongqun ZX, Mobley HL (2010) Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol https://doi.org/10.1155/2010/297505
Young KT, Davis LM, Dirita VJ (2007) Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5(9):665–679
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26(13):1608–1615
Acknowledgements
The authors wish to acknowledge the efforts of Dean, Faculty of Engineering and Technology, Rama University Uttar Pradesh, Kanpur for his experimental guidance during the course of experimental work. We are also grateful to supporting staff of Faculty of Engineering and Technology, Rama University Uttar Pradesh, Kanpur for providing necessary facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Gupta, N., Khan, F. & Kumar, A. Exploring Highly Antigenic Protein of Campylobacter jejuni for Designing Epitope Based Vaccine: Immunoinformatics Approach. Int J Pept Res Ther 25, 1159–1172 (2019). https://doi.org/10.1007/s10989-018-9764-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-018-9764-z