Abstract
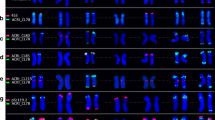
Fluorescence in situ hybridization was used to investigate the physical location of oligo-Am1 and (TTG)6 trinucleotide repeats in the metaphase chromosomes of seven diploid species (AA or CC genomes), seven tetraploid species (AABB or AACC genomes), and two hexaploid species (AACCDD genomes) belonging to the genus Avena. The oligo-Am1 probe produced signals that were particularly enriched on almost whole C-genome chromosomes, whereas the (TTG)6 probe was located in the pericentromeric (M), and, occasionally, their telomeric (T) chromosome regions, but showed low matching to C genome. All the species possessed (TTG)6 loci in M regions, and the CC, AABB, and AACCDD species also possessed (TTG)6 loci in T regions. The (TTG)6 signal number is constant in both the AA and CC species but slightly differs in signal intensity, whereas the (TTG)6 signal pattern shows wide diversity in the AABB, AACC and AACCDD species. The probe hybridization results provide key information that can be used in the physical assignment of genome sequences to chromosomes.




Similar content being viewed by others
Abbreviations
- DAPI:
-
4,6-Diamidino-2-phenylindole
- FAM:
-
6-Carboxyfluorescein
- TAMRM:
-
6-Carboxytetramethylrhodamine
- FISH:
-
Fluorescence in situ hybridization
- GISH:
-
Genomic in situ hybridization
- GBS:
-
Genotyping-by-sequencing
- M:
-
Pericentromic
- T:
-
Telomeric
References
Alicchio R, Aranci L, Conte L (1995) Restriction fragment length polymorphism based phylogenetic analysis of Avena L. Genome 38:1279–1284. https://doi.org/10.1139/g95-168
Badaeva ED, Shelukhina OY, Diederichsen A, Loskutov IG, Pukhalskiy VA (2010a) Comparative cytogenetic analysis of Avena macrostachya and diploid C-genome Avena species. Genome 53:125–137. https://doi.org/10.1139/g09-089
Badaeva ED, Shelukhina OY, Goryunova SV, Loskutov IG, Pukhalskiy VA (2010b) Phylogenetic relationships of tetraploid AB-genome Avena species evaluated by means of cytogenetic (C-banding and FISH) and RAPD analyses. J Bot 2010:1–13. https://doi.org/10.1155/2010/742307
Badaeva ED, Shelukhina OY, Dedkova OS, Loskutov IG, Pukhalskyi VA (2011) Comparative cytogenetic analysis of hexaploid Avena L. Species. Russ J Genet 47:691–702. https://doi.org/10.1134/s1022795411060068
Baum BR (1968) On some relationships between Avena sativa and A. fatua (Gramineae) as studied from Canadian material. Can J Bot 46:1013–1024. https://doi.org/10.1139/b68-135
Baum BR (1977) Oats wild and cultivated. A monograph of the genus Avena L. (Poaceae). Agriculture Canada. ISBN: 9780660005133
Bennett MD, Leitch IJ (2012) Plant DNA C-values database. Release 6.0. Dec 2012
Chaffin AS, Huang YF, Smith S, Bekele WA, Babiker E, Gnanesh BN, Foresman BJ, Blanchard SG, Jay JJ, Reid RW, Wight CP, Chao S, Oliver R, Islamovic E, Kolb FL, McCartney C, Mitchell Fetch JW, Beattie AD, Bjornstad A, Bonman JM, Langdon T, Howarth CJ, Brouwer CR, Jellen EN, Klos KE, Poland JA, Hsieh TF, Brown R, Jackson E, Schlueter JA, Tinker NA (2016) A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome 9(2):1–21. https://doi.org/10.3835/plantgenome2015.10.0102
Chen Q, Armstrong K (1994) Genomic in situ hybridization in Avena sativa. Genome 37:607–612 PMID:18470104
Chen Q, Armstrong K (1995) Characterization of a library from a single microdissected oat (Avena sativa L.) chromosome. Genome 38:706–714. http://www.ncbi.nlm.nih.gov/pubmed/18470104
Cheng DW, Armstrong KC, Drouin G, McElroy A, Fedak G, Molnar SD (2003) Isolation and identification of Triticeae chromosome 1 receptor-like kinase genes (lrk10) from diploid, tetraploid, and hexaploid species of the genus Avena. Genome 46:119–127. http://www.ncbi.nlm.nih.gov/pubmed/12669804
Craig IL, Murray BE, Rajhathy T (1974) Avena canariensis: morphological and electrophoretic polymorphism and relationship to the A. magna–A. murphyi complex and A. sterilis. Can J Genet Cytol 16:677–689. https://doi.org/10.1139/g74-074
Du P, Zhuang L, Wang Y, Yuan L, Wang Q, Wang D, Dawadondup, Tan L, Shen J, Xu H, Zhao H, Chu C, Qi Z (2017) Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60:93–103. http://www.ncbi.nlm.nih.gov/pubmed/27936984
Fabijanski S, Fedak G, Armstrong K, Altosaar I (1990) A repeated sequence probe for the c genome in Avena (oats). Theor Appl Genet 79:1–7. https://doi.org/10.1007/BF00223778
Fominaya A, Hueros G, Loarce Y, Ferrer E (1995) Chromosomal distribution of a repeated DNA sequence from C-genome heterochromatin and the identification of a new ribosomal DNA locus in the Avena genus. Genome 38:548–557. http://www.ncbi.nlm.nih.gov/pubmed/7557363
Fominaya A, Loarce Y, Montes A, Ferrer E (2017) Chromosomal distribution patterns of the (AC)10 microsatellite and other repetitive sequences, and their use in chromosome rearrangement analysis of species of the genus Avena. Genome 60:216–227. http://www.ncbi.nlm.nih.gov/pubmed/28156137
Gutierrez-Gonzalez JJ, Tu ZJ, Garvin DF (2013) Analysis and annotation of the hexaploid oat seed transcriptome. BMC Genome 14:471. https://doi.org/10.1186/1471-2164-14-471
Hao M, Luo J, Zhang L, Yuan Z, Yang Y, Wu M, Chen W, Zheng Y, Zhang H, Liu D (2013) Production of hexaploid triticale by a synthetic hexaploid wheat-rye hybrid method. Euphytica 193:347–357. https://doi.org/10.1007/s10681-013-0930-2
Hayasaki M, Morikawa T, Leggett JM (2001) Intraspecific variation of 18S-5.8S-26S rDNA sites revealed by FISH and RFLP in wild oat, Avena agadiriana. Genes Genet Syst 76:9–14. http://www.ncbi.nlm.nih.gov/pubmed/11376554
Huang YF, Poland JA, Wight CP, Jackson EW, Tinker NA (2014) Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PLOS ONE 9:e102448. http://www.ncbi.nlm.nih.gov/pubmed/25047601
Irigoyen ML, Loarce Y, Linares C, Ferrer E, Leggett M, Fominaya A (2001) Discrimination of the closely related a and b genomes in AABB tetraploid species of Avena. Theor Appl Genet 103:1160–1166. https://doi.org/10.1007/s001220100723
Irigoyen ML, Linares C, Ferrer E, Fominaya A (2002) Fluorescence in situ hybridization mapping of Avena sativa L. cv. Sun II and its monosomic lines using cloned repetitive DNA sequences. Genome 45:1230–1237. https://doi.org/10.1139/g02-076
Jellen EN, Bill BS (1996) C-banding variation in the Moroccan oat species Avena agadiriana (2n-2x = 28). Theor Appl Genet 92:726–732. https://doi.org/10.1007/BF00226095
Jellen EN, Gill BS, Cox TS (1994) Genomic in situ hybridization differentiates between a/d- and c-genome chromatin and detects intergenomic translocations in polyploid oat species (genus Avena). Genome 37:613–618. https://doi.org/10.1139/g94-087
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559. https://doi.org/10.1073/pnas.0403659101
Katsiotis A, Schmidt T, Heslop-Harrison JS (1996) Chromosomal and genomic organization ofty1-copia-like retrotransposon sequences in the genus Avena. Genome 39:410–417. https://doi.org/10.1139/g96-052
Kellogg EA, Bennetzen JL (2004) The evolution of nuclear genome structure in seed plants. Am J Bot 91:1709–1725. https://doi.org/10.3732/ajb.91.10.1709
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137. https://doi.org/10.1139/gen-2013-0003
Ladizinsky G (1998) A new species of Avena from sicily, possibly the tetraploid progenitor of hexaploid oats. Genet Resour Crop Evol 45:263–269. https://doi.org/10.1023/a:1008657530466
Loskutov IG, Rines HW (2011) Avena. In: Kole C (ed) Wild crop relatives: genomic and breeding resources. Springer, Heidelberg, pp 109–183. https://doi.org/10.1007/978-3-642-14228-4_3
Luo X, Zhang H, Kang H, Fan X, Wang Y, Sha L, Zhou Y (2014) Exploring the origin of the D genome of oat by fluorescence in situ hybridization. Genome 57:469–472. https://doi.org/10.1139/gen-2014-0048
Luo X, Tinker NA, Zhang H, Wight CP, Kang H, Fan X, Wang Y, Sha L, Zhou Y (2015) Centromeric position and genomic allocation of a repetitive sequence isolated from chromosome 18D of hexaploid oat, Avena sativa L. Genet Resour Crop Evol 62:1–4. https://doi.org/10.1007/s10722-014-0170-x
Luo X, Tinker NA, Zhou Y, Wight PC, Wan W, Chen L, Peng Y (2018) Genomic relationships among sixteen Avena taxa based on (ACT)6 trinucleotide repeat FISH. Genome 61:63–70. https://doi.org/10.1139/gen-2017-0132
Nikoloudakis N, Katsiotis A (2008) The origin of the C-genome and cytoplasm of Avena polyploids. Theor Appl Genet 117:273–281. https://doi.org/10.1007/s00122-008-0772-9
Pedersen C, Rasmussen SK, Linde-Laursen I (1996) Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39:93–104. https://doi.org/10.1139/g96-013
Rajhathy T, Thomas H (1974) Cytogenetics of oats (Avena L.). Genetics Society of Canada, Ottawa. Miscellaneous Publ Genet Soc Can, p 90. https://www.researchgate.net/publication/37875550_Cytogenetics_of_oats_Avena_L
Rodrigues J, Viegas W, Silva M (2017) 45S rDNA external transcribed spacer organization reveals new phylogenetic relationships in Avena genus. PLoS ONE 12:e0176170. https://doi.org/10.1371/journal.pone.0176170
Sanz MJ, Loarce Y, Ferrer E, Fominaya A (2012) Use of tyramide-fluorescence in situ hybridization and chromosome microdissection for ascertaining homology relationships and chromosome linkage group associations in oats. Cytogenet Genome 36:145–156. https://doi.org/10.1159/000335641
Thomas H (1989) New species of Avena. In: Proceedings of the 3rd international oat conference, Lund, Sweden, pp 18–23. https://doi.org/10.1017/s1477200007002447
Tomás D, Rodrigues J, Varela A, Veloso MM, Viegas W, Silva M (2016) Use of repetitive sequences for molecular and cytogenetic characterization of Avena species from Portugal. Int J Mol Sci 7:203. https://doi.org/10.3390/ijms17020203
Yan H, Bekele WA, Wight CP, Peng Y, Langdon T, Latta RG, Fu YB, Diederichsen A, Howarth CJ, Jellen EN, Boyle B, Wei Y, Tinker NA (2016a) High-density marker profiling confirms ancestral genomes of Avena species and identifies D-genome chromosomes of hexaploid oat. Theor Appl Genet 129:2133–2149. http://www.ncbi.nlm.nih.gov/pubmed/27522358
Yan H, Martin SL, Bekele WA, Latta RG, Diederichsen A, Peng Y, Tinker NA (2016b) Genome size variation in the genus Avena. Genome 59:209–220. http://www.ncbi.nlm.nih.gov/pubmed/26881940
Yang Q, Hanson L, Bennett MD, Leitch IJ (1999) Genome structure and evolution in the allohexaploid weed Avena fatua L. (Poaceae). Genome 42:512–518. https://doi.org/10.1139/g98-154
Zhao LB, Ning SZ, Yu JJ, Hao M, Zhang LQ, Yuan ZW, Zheng YL, Liu DC (2016) Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed Sci 66:522–529. https://doi.org/10.1270/jsbbs.16011
Acknowledgements
This study was supported by the Natural Science Foundation of China (31500993). The authors greatly appreciate the American National Plant Germplasm System (Pullman, WA, USA) and Plant Gene Resources of Canada (Saskatoon, SK, Canada) for providing the material for the investigation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Luo, X., Tinker, N.A., Zhou, Y. et al. Chromosomal distributions of oligo-Am1 and (TTG)6 trinucleotide and their utilization in genome association analysis of sixteen Avena species. Genet Resour Crop Evol 65, 1625–1635 (2018). https://doi.org/10.1007/s10722-018-0639-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0639-0