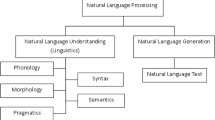
Abstract
In line with the current trajectory of healthcare reform, significant emphasis has been placed on improving the utilization of data collected during a clinical encounter. Although the structured fields of electronic health records have provided a convenient foundation on which to begin such efforts, it was well understood that a substantial portion of relevant information is confined in the free-text narratives documenting care. Unfortunately, extracting meaningful information from such narratives is a non-trivial task, traditionally requiring significant manual effort. Today, computational approaches from a field known as Natural Language Processing (NLP) are poised to make a transformational impact in the analysis and utilization of these documents across healthcare practice and research, particularly in procedure-heavy sub-disciplines such as gastroenterology (GI). As such, this manuscript provides a clinically focused review of NLP systems in GI practice. It begins with a detailed synopsis around the state of NLP techniques, presenting state-of-the-art methods and typical use cases in both clinical settings and across other domains. Next, it will present a robust literature review around current applications of NLP within four prominent areas of gastroenterology including endoscopy, inflammatory bowel disease, pancreaticobiliary, and liver diseases. Finally, it concludes with a discussion of open problems and future opportunities of this technology in the field of gastroenterology and health care as a whole.
Similar content being viewed by others
References
Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;17:128–144.
Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42:760–772.
Leaman R, Khare R, Lu Z. Challenges in clinical natural language processing for automated disorder normalization. J Biomed Inform. 2015;57:28–37.
Velupillai S, Suominen H, Liakata M, et al. Using clinical Natural Language Processing for health outcomes research: overview and actionable suggestions for future advances. J Biomed Inform. 2018;88:11–19.
Névéol A, Dalianis H, Velupillai S, Savova G, Zweigenbaum P. Clinical natural language processing in languages other than english: opportunities and challenges. J Biomed Semant. 2018;9:12.
Carrell DS, Schoen RE, Leffler DA, et al. Challenges in adapting existing clinical natural language processing systems to multiple, diverse health care settings. J Am Med Inform Assoc JAMIA. 2017;24:986–991.
Zozus MN, Pieper C, Johnson CM, et al. Factors affecting accuracy of data abstracted from medical records. PloS One. 2015;10:e0138649.
Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform. 2018;77:34–49.
Luque C, Luna JM, Luque M, Ventura S. An advanced review on text mining in medicine. WIREs Data Min Knowl Discov. 2019;9:e1302.
Simpson MS, Demner-Fushman D. Biomedical text mining: a survey of recent progress. In: Aggarwal CC, Zhai C, eds. Mining Text Data. Boston: Springer; 2012:465–517.
Belskaja IK. Machine translation of languages. Research. 1957;10:383–389.
Manning CD, Raghavan P, Schütze H. Introduction to Information Retrieval. Cambridge: Cambridge University Press; 2008.
Jurafsky D, Martin JH. Speech and language processing: an introduction to natural language processing, computational linguistics, and speech recognition. Upper Saddle River: Pearson Prentice Hall; 2009.
Calijorne Soares MA, Parreiras FS. A literature review on question answering techniques, paradigms and systems. J King Saud Univ Comput Inf Sci. 2018. https://doi.org/10.1016/j.jksuci.2018.08.005.
Lloret E. Palomar M Text summarisation in progress: a literature review. Artif Intell Rev. 2012;37:1–41.
Gentzkow M, Kelly B, Taddy M. Text as data. J Econ Lit.. 2019;57:535–574.
Crawford M, Khoshgoftaar TM, Prusa JD, Richter AN, Al Najada H. Survey of review spam detection using machine learning techniques. J Big Data. 2015;2:23.
Liddy ED. Natural language processing. In: Drake MA, ed. Encyclopedia of Library and Information Science, 2nd Ed. New York: Marcel Decker, Inc.; 2001.
Blei DM. Probabilistic topic models. Commun ACM. 2012;55:77–84.
Manning CD. Part-of-speech tagging from 97% to 100%: Is it time for some linguistics? In: Gelbukh AF, ed. Computational Linguistics and Intelligent Text Processing. Berlin: Springer; 2011:171–189.
Ferraro JP, Daumé H, DuVall SL, Chapman WW, Harkema H, Haug PJ. Improving performance of natural language processing part-of-speech tagging on clinical narratives through domain adaptation. J Am Med Inform Assoc JAMIA. 2013;20:931–939.
Goyal A, Gupta V, Kumar M. Recent Named Entity Recognition and Classification techniques: a systematic review. Comput Sci Rev. 2018;29:21–43.
Kübler S, McDonald R, Nivre J. Dependency parsing. Synth Lect Hum Lang Technol. 2008;2:1–127.
Shen W, Wang J, Han J. Entity linking with a knowledge base: issues, techniques, and solutions. IEEE Trans Knowl Data Eng. 2015;27:443–460.
Giatsoglou M, Vozalis MG, Diamantaras K, Vakali A, Sarigiannidis G, Chatzisavvas KCh. Sentiment analysis leveraging emotions and word embeddings. Expert Syst Appl. 2017;69:214–224.
Mitra B, Craswell N. Neural text embeddings for information retrieval. In: Proceedings of the Tenth ACM International Conference on Web Search and Data Mining. Association for Computing Machinery, New York, NY, USA; 2017:813–814.
Bengio S, Heigold G. Word embeddings for speech recognition. In: Interspeech; 2014:1053–1057.
Le Q, Mikolov T. Distributed representations of sentences and documents. In: Proceedings of the 31st International Conference on International Conference on Machine Learning; volume 32; 2014.
Sennrich R, Haddow B, Birch A. Neural machine translation of rare words with subword units. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Berlin, Germany: Association for Computational Linguistics; 2016:1715–1725.
Kim Y, Jernite Y, Sontag D, Rush AM. Character-aware neural language models. CoRR, 2015.
Wieting J, Bansal M, Gimpel K, Livescu K. Charagram: embedding words and sentences via character n-grams. In: EMNLP. 2016.
Assale M, Dui LG, Cina A, Seveso A, Cabitza F. The revival of the notes field: leveraging the unstructured content in electronic health records. Front Med. 2019. https://doi.org/10.3389/fmed.2019.00066.
Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc JAMIA. 2011;18:544–551.
Cai T, Giannopoulos AA, Yu S, et al. Natural language processing technologies in radiology research and clinical applications. Radiogr Rev Publ Radiol Soc N Am Inc. 2016;36:176–191.
Yim W-W, Yetisgen M, Harris WP, Kwan SW. Natural language processing in oncology: a review. JAMA Oncol. 2016;2:797–804.
Pons E, Braun LMM, Hunink MGM, Kors JA. Natural language processing in radiology: a systematic review. Radiology. 2016;279:329–343.
Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306:848–855.
Vo E, Davila JA, Hou J, et al. Differentiation of ileostomy from colostomy procedures: assessing the accuracy of current procedural terminology codes and the utility of natural language processing. Surgery. 2013;154:411–417.
Elkin PL, Froehling DA, Wahner-Roedler DL, Brown SH, Bailey KR. Comparison of natural language processing biosurveillance methods for identifying influenza from encounter notes. Ann Intern Med. 2012;156:11–18.
Calvo RA, Milne DN, Hussain MS, Christensen H. Natural language processing in mental health applications using non-clinical texts. Nat Lang Eng. 2017;23:649–685.
Conway M, O’Connor D. Social media, big data, and mental health: current advances and ethical implications. Curr Opin Psychol. 2016;9:77–82.
Sun W, Cai Z, Li Y, Liu, F, Fang S, Wang G. Data processing and text mining technologies on electronic medical records: a review. J Healthc Eng. 2018;2018:4302425.
Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:267–270.
McCray AT. An upper-level ontology for the biomedical domain. Int J Genom. 2003;4:80–84.
Zhang Y, Chen Q, Yang Z, Lin H, Lu Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci Data. 2019. https://doi.org/10.1038/s41597-019-0055-0.
Gero Z, Ho J. PMCVec: distributed phrase representation for biomedical text processing. J Biomed Inform. 2019;3:100047.
Bai T, Chanda AK, Egleston BL, Vucetic S. EHR phenotyping via jointly embedding medical concepts and words into a unified vector space. BMC Med Inform Decis Mak. 2018;18:123.
Liu K, Hogan WR, Crowley RS. Natural Language Processing methods and systems for biomedical ontology learning. J Biomed Inform. 2011;44:163–179.
Feczko PJ, Ackerman LV, Halpert RD, Simms SM. A computer-based gastrointestinal information management system. Radiology. 1984;152:297–300.
Rex DK, Schoenfeld PS, Cohen J, et al. Quality indicators for colonoscopy. Am J Gastroenterol. 2015;110:72–90.
Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298–1306.
Barclay RL, Vicari JJ, Doughty AS, Johanson JF, Greenlaw RL. Colonoscopic withdrawal times and adenoma detection during screening colonoscopy. N Engl J Med. 2006;355:2533–2541.
Adler A, Wegscheider K, Lieberman D, et al. Factors determining the quality of screening colonoscopy: a prospective study on adenoma detection rates, from 12,134 examinations (Berlin colonoscopy project 3, BECOP-3). Gut. 2013;62:236–241.
Rex DK. Who is the best colonoscopist? Gastrointest Endosc. 2007;65:145–150.
Pike IM. Quality improvement in gastroenterology: a US perspective. Nat Clin Pract Gastroenterol Hepatol. 2008;5:550–551.
Imler TD, Morea J, Kahi C, et al. Multi-center colonoscopy quality measurement utilizing natural language processing. Am J Gastroenterol. 2015;110:543–552.
Imler TD, Morea J, Kahi C, Imperiale TF. Natural language processing accurately categorizes findings from colonoscopy and pathology reports. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2013;11:689–694.
Mehrotra A, Dellon ES, Schoen RE, et al. Applying a natural language processing tool to electronic health records to assess performance on colonoscopy quality measures. Gastrointest Endosc. 2012;75:1233–1239.
Harkema H, Chapman WW, Saul M, Dellon ES, Schoen RE, Mehrotra A. Developing a natural language processing application for measuring the quality of colonoscopy procedures. J Am Med Inform Assoc JAMIA. 2011;18:i150–i156.
Gawron AJ, Thompson WK, Keswani RN, Rasmussen LV, Kho AN. Anatomic and advanced adenoma detection rates as quality metrics determined via natural language processing. Am J Gastroenterol. 2014;109:1844–1849.
Lee JK, Jensen CD, Levin TR, et al. Accurate identification of colonoscopy quality and polyp findings using natural language processing. J Clin Gastroenterol. 2019;53:e25–e30.
Raju GS, Lum PJ, Slack RS, et al. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointest Endosc. 2015;82:512–519.
Deutsch JC. Colonoscopy quality, quality measures, and a natural language processing tool for electronic health records. Gastrointest Endosc. 2012;75:1240–1242.
Marcondes FO, Gourevitch RA, Schoen RE, Crockett SD, Morris M, Mehrotra A. Adenoma detection rate falls at the end of the day in a large multi-site sample. Dig Dis Sci. 2018;63:856–859. https://doi.org/10.1007/s10620-018-4947-1.
Patel VD, Thompson WK, Lapin BR, Goldstein JL, Yen EF. Screening colonoscopy withdrawal time threshold for adequate proximal serrated polyp detection rate. Dig Dis Sci. 2018;63:3084–3090. https://doi.org/10.1007/s10620-018-5187-0.
Abdul-Baki H, Schoen RE, Dean K, et al. Public reporting of colonoscopy quality is associated with an increase in endoscopist adenoma detection rate. Gastrointest Endosc. 2015;82:676–682.
Mehrotra A, Morris M, Gourevitch RA, et al. Physician characteristics associated with higher adenoma detection rate. Gastrointest Endosc. 2018;87:778–786.
Crockett SD, Gourevitch RA, Morris M, et al. Endoscopist factors that influence serrated polyp detection: a multicenter study. Endoscopy. 2018;50:984–992.
Gourevitch RA, Rose S, Crockett SD, et al. Variation in pathologist classification of colorectal adenomas and serrated polyps. Am J Gastroenterol. 2018;113:431–439.
Hong SN, Son HJ, Choi SK, et al. A prediction model for advanced colorectal neoplasia in an asymptomatic screening population. PloS One. 2017;12:e0181040.
Blumenthal DM, Singal G, Mangla SS, Macklin EA, Chung DC. Predicting non-adherence with outpatient colonoscopy using a novel electronic tool that measures prior non-adherence. J Gen Intern Med. 2015;30:724–731.
Thrift AP, Natarajan Y, Mansour NM, et al. Tu1117—using natural language processing to accurately identify dysplasia in pathology reports for patients with Barrett’s Esophagus. Gastroenterology. 2018;154:896–897.
Hou JK, Soysal E, Moon S, et al. Su1815 natural language processing accurately identifies colorectal dysplasia in a national cohort of veterans with inflammatory bowel disease. Gastroenterology. 2016;150:S560–S561.
Ananthakrishnan AN, Cai T, Cheng S-C, et al. Tu1276 improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing—a novel informatics approach. Gastroenterology. 2012;142:S-791.
Cai T, Lin T-C, Bond A, et al. The association between arthralgia and Vedolizumab using natural language processing. Inflamm Bowel Dis. 2018;24:2242–2246.
Hou JK, Chang M, Nguyen T, et al. Automated identification of surveillance colonoscopy in inflammatory bowel disease using natural language processing. Dig Dis Sci. 2013;58:936–941. https://doi.org/10.1007/s10620-012-2433-8.
ASGE Standards of Practice Committee, Anderson MA, Fisher L, et al. Complications of ERCP. Gastrointest Endosc. 2012;75:467–473.
Imler TD, Sherman S, Imperiale TF, et al. Provider-specific quality measurement for ERCP using natural language processing. Gastrointest Endosc. 2018;87:164–173.
Al-Haddad MA, Friedlin J, Kesterson J, Waters JA, Aguilar-Saavedra JR, Schmidt CM. Natural language processing for the development of a clinical registry: a validation study in intraductal papillary mucinous neoplasms. HPB. 2010;12:688–695.
Mehrabi S, Schmidt CM, Waters JA, et al. An efficient pancreatic cyst identification methodology using natural language processing. Stud Health Technol Inform. 2013;192:822–826.
Chang EK, Yu CY, Clarke R, et al. Mo1050 using an automated diagnostic algorithm that utilizes electronic health records and natural language processing to define a population with cirrhosis. Gastroenterology.. 2015;148:S-1074.
Kung R, Ma A, Dever JB, et al. Mo1043 a natural language processing algorithm for identification of patients with cirrhosis from electronic medical records. Gastroenterology. 2015;148:1071–1072.
Corey KE, Kartoun U, Zheng H, Shaw SY. Development and validation of an algorithm to identify nonalcoholic fatty liver disease in the electronic medical record. Dig Dis Sci. 2016;61:913–919. https://doi.org/10.1007/s10620-015-3952-x.
Sada Y, Hou J, Richardson P, El-Serag H, Davila J. Validation of case finding algorithms for hepatocellular cancer from administrative data and electronic health records using natural language processing. Med Care. 2016;54:e9–e14.
Koola JD, Davis SE, Al-Nimri O, et al. Development of an automated phenotyping algorithm for hepatorenal syndrome. J Biomed Inform. 2018;80:87–95.
Imler TD, Ring N, Crabb DW. Su1403 medical, social, and legal risks to predict alcoholic liver disease using natural language processing and advanced analytics. Gastroenterology. 2015;148:S-499.
Jha AK. The promise of electronic records: around the corner or down the road? JAMA. 2011;306:880–881.
Imler TD, Morea J, Imperiale TF. Clinical decision support with natural language processing facilitates determination of colonoscopy surveillance intervals. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2014;12:1130–1136.
Bhat S, Zahorian T, Robert R, Farraye FA. Advocating for patients with inflammatory bowel disease: how to navigate the prior authorization process. Inflamm Bowel Dis. 2019;25:1621–1628.
Walsh SH. The clinician’s perspective on electronic health records and how they can affect patient care. BMJ. 2004;328:1184–1187.
Coiera E. When conversation is better than computation. J Am Med Inform Assoc JAMIA. 2000;7:277–286.
Maybury M. Advances in Automatic Text Summarization. New York: MIT Press; 1999.
Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med. 1999;74:890–895.
Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility Pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak. 2015. https://doi.org/10.1186/s12911-015-0149-3.
Trugenberger CA, Wälti C, Peregrim D, Sharp ME, Bureeva S. Discovery of novel biomarkers and phenotypes by semantic technologies. BMC Bioinform. 2013;14:51.
Coppersmith G, Leary R, Crutchley P, Fine A. Natural language processing of social media as screening for suicide risk. Biomed Inform Insights. 2018. https://doi.org/10.1177/1178222618792860.
McCoy AB, Wright A, Eysenbach G, et al. State of the art in clinical informatics: evidence and examples. Yearb Med Inform. 2013;8:13–19.
Zheng X, Feng J, Chen Y, Peng H, Zhang W. Learning context—specific word/character embeddings. In: Proceedings of AAAI Conference on Artificial Intelligence; 2017:3393–3399.
Li P, Bing L, Lam W, Li H, Liao Y. Reader-aware multi-document summarization via sparse coding. In: IJCAI; 2015.
Forbush TB, Gundlapalli AV, Palmer MN, et al. Sitting on Pins and needles: characterization of symptom descriptions in clinical notes. AMIA Summits Transl Sci Proc. 2013;2013:67–71.
Koleck TA, Dreisbach C, Bourne PE, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc. 2019;26:364–379.
Chapman WW. Closing the gap between NLP research and clinical practice. Methods Inf Med. 2010;49:317–319.
Liberati EG, Ruggiero F, Galuppo L, et al. What hinders the uptake of computerized decision support systems in hospitals? A qualitative study and framework for implementation. Implement Sci. 2017;12:113.
Horsky J, Schiff GD, Johnston D, Mercincavage L, Bell D, Middleton B. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J Biomed Inform. 2012;45:1202–1216.
Acknowledgments
We would like to thank An-Lin Cheng, PhD from the department of Bioinformatics at the University of Missouri-Kansas City for her assistance and expertise in developing this manuscript.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This statement is to certify that all authors have no conflict of interest related to the current manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nehme, F., Feldman, K. Evolving Role and Future Directions of Natural Language Processing in Gastroenterology. Dig Dis Sci 66, 29–40 (2021). https://doi.org/10.1007/s10620-020-06156-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06156-y