Abstract
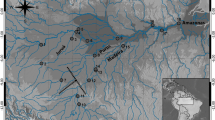
The Mekong River is a vital fisheries resource supporting millions of people in mainland Southeast Asia. However, numerous threats have the potential to negatively impact fish populations in this region including overfishing, pollution, climate change and increased urban, agriculture and upstream hydropower development. Although a few studies have examined the population genetic structure of fishes within the upper Mekong River, no known studies have explored that of fishes within the Mekong Delta (MD). Here, we examine the population structure of an important food fish within the MD, Polynemus melanochir, using a panel of 1735 single nucleotide polymorphisms (SNPs) generated by restriction site-associated DNA (RAD) sequencing across eight locations on the Tien (Mekong) and Hau (Bassac) Rivers in Vietnam. Pairwise FST values, principal component analysis and Structure analysis all indicate high levels of gene flow among the sites sampled across the MD. In contrast to the lack of genetic structure, high levels of relatedness were found, including 26 putatively related pairs, as well as an effective population size (Ne) of less than 500 across the MD. While panmixia indicates that fragmentation of this population is not presently an important threat, a low Ne estimate suggests this species may not be resilient to long-term environmental changes in the MD. The reliance on P. melanochir as a food resource may be contingent on management and mitigation of low effective population sizes.



Similar content being viewed by others
References
Adamson EA, Hurwood DA, Baker AM, Mather PB (2009) Population subdivision in Siamese mud carp Henicorhynchus siamensis in the Mekong River basin: implications for management. J Fish Biol 75(6):1371–1392
Allen DJ, Smith KG, Darwall WR (2012) The status and distribution of freshwater biodiversity in Indo-Burma. IUCN, Gland
Alliance Delta (2011) Mekong Delta water resources assessment studies Vietnam-Netherlands Mekong Delta Masterplan project. Deltares, Delft
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9:323
Arias ME, Cochrane T, Norton D, Killeen TJ, Khon P (2013) The flood pulse as the underlying driver of vegetation in the largest wetland and fishery of the Mekong basin. Ambio 42(7):864–876
Attard CR, Beheregaray LB, Möller LM (2018) Genotyping-by-sequencing for estimating relatedness in nonmodel organisms: avoiding the trap of precise bias. Mol Ecol Resour 18(3):381–390
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B 263:1619–1626
Beerli PE (2009) How to use MIGRATE or why are Markov chain Monte Carlo programs difficult to use. In: Bertorelle G, Bruford MW, Hauffe HC, Rizzoli A, Vernesi (eds) Population genetics for animal conservation. The Cambridge University Press, Cambridge, pp. 42–79
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population size in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98:4563–4568
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57(1):289–300
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Campbell, IC (2012). Biodiversity of the Mekong Delta. In: The Mekong Delta system. Springer, Dordrecht, pp. 293–313
Chenoweth SF, Hughes JM (2003) Oceanic interchange and nonequilibrium population structure in the estuarine dependent Indo Pacific tasselfish Polynemus sheridani. Mol Ecol 12(9):2387–2397
Chong Z, Ruan J, Wu CI (2012) Rainbow: an integrated tool for efficient clustering and assembling RAD-seq reads. Bioinformatics 28(21):2732–2737
Cruz RV, Harasawa H, Lal M, Wu S, Anokhin Y, Punsalmaa B, Honda Y, Jafari M, Li C, Huu Ninh N (2007) Asia in Climate change 2007: impacts, adaptation and vulnerability. In: Parry ML, Canziani OF, Palutikof JP, van der Linden PJ and Hanson CE (eds) Contribution of Working Group II to the fourth assessment report of the Intergovernmental Panel on Climate Change, pp. 469–506. Cambridge University Press, Cambridge
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158
Dasgupta S, Laplante B, Meisner C, Wheeler D, Yan J (2009) The impact of sea level rise on developing countries: a comparative analysis. Clim Chang 93:379–388
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (N e) from genetic data. Mol Eco Resour 14:209–214
Dutta D, Alam J, Umeda K, Hayashi M, Hironaka S (2007) A two-dimensional hydrodynamic model for flood inundation simulation: a case study in the lower Mekong river basin. Hydrol Process 21:1223–1237
Earl DA, von Holdt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14(8):2611–2620
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180(2):977–993
Frankham R, Bradshaw CJ, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Franklin IR, Allendorf FW, Jamieson IG (2014) The 50, 500 rule is still valid: reply to Frankham et al. Biol Conserv 176:284–285
Fu F, Tarnita CE, Christakis NA, Wang L, Rand DG, Nowak MA (2012) Evolution of in-group favoritism. Sci Rep 2:1–6
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv Preprint http://arxiv.org/abs/1207.3907
Gido KB, Whitney JE, Perkin JS, Turner TF (2016) Fragmentation, connectivity and fish species persistence in freshwater ecosystems. Conserv Fresh Fish 292–323
Goldberg CS, Waits LP (2010) Quantification and reduction of bias from sampling larvae to infer population and landscape genetic structure. Mol Ecol Resour 10:304–313
Gugliotta M, Saito Y, Nguyen VL, Ta TK, Nakashima R, Tamura T, Uehara K, Katsuki K, Yamaoto S (2017) Process regime, salinity, morphological, and sedimentary trends along the fluvial to marine transition zone o fthe mixed-energy Mekong River dealta, Vietnam. Cont Shelf Res 147:7–26
Hak D, Nadaoka K, Bernado LP, Le Phu V, Quan NH, Toan TQ, Trung NH, Van Ni D, Van PD (2016) Spatio-temporal variations of sea level around the Mekong Delta: their causes and consequences on the coastal environment. Hydrol Res Lett 10:60–66
Hurwood DA, Adamson EA, Mather PB (2008) Evidence for strong genetic structure in a regionally important, highly vagile cyprinid (Henicorhynchus lobatus) in the Mekong River Basin. Ecol Freshw Fish 17(2):273–283
James KR, Cant B, Ryan T (2003) Responses of freshwater biota to rising salinity levels and implications for saline water management: a review. Aust J Bot 51:703–713
Jamieson IG, Allendorf FW (2012) How does the 50/500 rule apply to MVPs? Trends Ecol Evol 27(10):578–584
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Junk W, Bayley PB, Sparks RE (1989) The flood pulse concept in river floodplain systems. In: Dodge DP (ed) Proceedings of the international large river symposium (LARS). Can B Fish Aquat Sci 106:110–127
Koehn JD, Hobday AJ, Pratchett MS, Gillanders BM (2011) Climate change and Australian marine and freshwater environments, fishes and fisheries: synthesis and options for adaptation. Mar Freshw Res 62:1148–1164
Kottelat M (2013) The fishes of the inland waters of Southeast Asia: a catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles B Zool, Supplement 27:1–663
Le AT, Chu TH, Miller F, Bach TS (2007a) Flood and salinity management in the Mekong Delta, Vietnam. In: Be, TT, Sinh BT, Miller F (eds) Challenges to sustainable development in the Mekong Delta: regional and national policy issues and research needs. Bangkok, Thailand
Le TV, Nguyen HN, Wolanski E, Tran TC, Haruyama S (2007) The combined impact on the flooding in Vietnam's Mekong River delta of local man-made structures, sea level rise, and dams upstream in the river catchment. Estuar Coast Shelf S 71:110–116
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint http://arxiv.org/abs/1303.3997
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Milligan BG (2003) Maximum-likelihood estimation of relatedness. Genetics 163:1153–1167
Motomura H (2004) FAO species catalogue. Threadfins of the world (family Polynemidae). An annotated and illustrated catalogue of polynemid species known to date. FAO, Rome
Motomura H, Sabaj MH (2002) A new subspecies, Polynemus melanochir dulcis, from Tonle Sap Lake, Cambodia, and redescription of P. m. melanochir Valenciennes in Cuvier and Valenciennes, 1831 with designation of a neotype. Ichthyol Res 49:181–190
Motomura H, Tsukawaki S (2006) New species of the threadfin genus Polynemus (Teleostei: Polynemidae) from the Mekong River basin, Vietnam, with comments on the Mekong species of Polynemus. Raffles B Zool 54:459–464
Nahar A, Siddik MA, Alam MA, Chaklader MR (2015) Population genetic structure of paradise threadfin Polynemus paradiseus (Linnaeus, 1758) revealed by allozyme marker. Int J Zool Res 1(2):48–56
Nguyen N (2017) Historic drought and salinity intrusion in the Mekong Delta in 2016: lessons learned and response solutions. Vietnam Sci Technol 1:93–96
Nguyen AD, Savenije HH (2006) Salt intrusion in multi-channel estuaries: a case study in the Mekong Delta. Vietnam. Hydrol Earth Syst Sc Discuss 10:743–754
Nguyen TT, Sunnucks P (2012) Strong population genetic structure and its management implications in the mud carp Cirrhinus molitorella, an indigenous freshwater species subject to an aquaculture and culture-based fishery. J Fish Biol 80(3):651–668
Nguyen LA, Verreth JA, Leemans HB, Bosma RH, De Silva S (2016) A decision tree analysis to support potential climate change adaptations of striped catfish (Pangasianodon hypophthalmus Sauvage) farming in the Mekong Delta, Vietnam. Tropicultura 34(Special):105–15
Noh S, Choi M, Kim E, Dan NP, Thanh BX, Van Ha NT, Sthiannopkao S, Han S (2013) Influence of salinity intrusion on the speciation and partitioning of mercury in the Mekong River Delta. Geochim Cosmochim Acta 106:379–390
O'Leary SJ, Puritz JB, Willis SC, Hollenbeck CM, Portnoy DS (2018) These aren’t the loci you’e looking for: principles of effective SNP filtering for molecular ecologists. Mol Ecol 27(16):3193–3206
Pew J, Muir PH, Wang J, Frasier TR (2015) Related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol Ecol Resour 15:557–561
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Puritz JB, Hollenbeck CM, Gold JR (2014) dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ 2:e431
Ritland K (1996) Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67(2):175–185
Samarasin P, Shuter BJ, Wright SI, Rodd FH (2016) The problem of estimating recent genetic connectivity in a changing world. Conserv Biol 31(1):126–135
Scribner KT, Lowe WH, Landguth E, Luikart G, Infante DM, Whelan GE, Muhlfeld CC (2016) Applications of genetic data to improve management and conservation of river fishes and their habitats. Fisheries 41(4):174–188
Smajgl A, Toan TQ, Nhan DK, Ward J, Trung NH, Tri LQ, Tri VP, Vu PT (2015) Responding to rising sea levels in the Mekong Delta. Nat Clim Chang 5:167
So N, Van Houdt JK, Volckaert FA (2006) Genetic diversity and population history of the migratory catfishes Pangasianodon hypophthalmus and Pangasius bocourti in the Cambodian Mekong River. Fish Sci 72(3):469–476
Sun X, Xu D, Lou B, Zhang T, Xin J, Guo Y, Ma S (2013) Genetic diversity and population structure of Eleutheronema rhadinum in the East and South China Seas revealed in mitochondrial COI sequences. Chin J Oceanol Limn 31(6):1276–1283
Ta TK, Nguyen VL, Tateishi M, Kobayashi I, Tanabe S, Saito Y (2002) Holocene delta evolution and sediment discharge of the Mekong River, southern Vietnam. Quaternary Sci Rev 21:1807–1819
Takagi AP, Ishikawa S, Nao T, Song SL, Hort S, Thammavong K, Saphakdy B, Phomsouvanhm A, Nishida M, Kurokura H (2010) Genetic differentiation and distribution routes of the bronze featherback Notopterus notopterus (Osteoglossiformes: Notopteridae) in Indochina. Biol J Linn Soc 101(3):575–582
Takagi AP, Ishikawa S, Nao T, Limsong S, Hort S, Thammavong K, Saphakdy B, Phomsouvanhm A, Nishida M, Kurokura H (2011) Population structure of the climbing perch, Anabas testudineus, in the lower Mekong River basin. Fish Manag Ecol 18(2):145–153
Toonen RJ, Puritz JB, Forsman ZH, Whitney JL, Fernandez-Silva I, Andrews KR, Bird CE (2013) ezRAD: a simplified method for genomic genotyping in non-model organisms. PeerJ 19(1):e203
Vo, KT (2012) Hydrology and hydraulic infrastructure systems in the Mekong Delta, Vietnam. In: The Mekong Delta system. Springer, Dordrecht, pp. 49–81
Wang J (2007) Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet Res 89(03):135–153
Waples RS, Anderson EC (2017) Purging putative siblings from population genetic data sets: a cautionary view. Mol Ecol 26(5):1211–1224
Acknowledgements
We thank the many people from the communities of the Mekong River Delta who helped us collect tissues from fish markets and team members of Biodiversity and Conservation, Institute for Biotechnology and Environment, Nha Trang University for project support. We are very grateful to Christopher Bird and Sharon Furiness Magnuson who provided help and advice in sequencing and data analysis. We thank Min Dong, Wirawan Purwanto, Terry Stilwell and John Pratt of the Old Dominion University High Performance Computing Facility for computational assistance. This project was funded by the United States Agency for International Development supported Partnerships for Enhanced Research Project 2–7 under USAID Cooperative Agreement AID-OAA-A-11-00012 and project OISE-0730256 sponsored by the United States National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dang, B.T., Vu, Q.H.D., Biesack, E.E. et al. Population genomics of the peripheral freshwater fish Polynemus melanochir (Perciformes, Polynemidae) in a changing Mekong Delta. Conserv Genet 20, 961–972 (2019). https://doi.org/10.1007/s10592-019-01189-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01189-x