Abstract
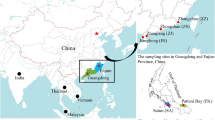
Eleutheronema rhadinum is a potential commercial fisheries species and is subject to intense exploitation in China. Knowledge on the population structure of E. rhadinum in Chinese coastal waters, which is important for sustainable exploitation and proper resource management, is lacking. In the present study, the genetic diversity and population structure of E. rhadinum were evaluated using a 564-base pair fragment of the mitochondrial cytochrome c oxidase subunit I (COI) gene. A total of 76 specimens were collected from three localities around the East (Qidong and Zhoushan) and South China Seas (Zhuhai). Among these individuals, nine polymorphic sites were detected and 11 distinct haplotypes were defined. High levels of haplotype diversity (h =0.759±0.035) and low levels of nucleotide diversity (π= 0.001 98±0.003 26) were observed in these populations. Hierarchical analysis of molecular variance (AMOVA) indicated that 96.72% of the genetic variation occurred within the populations, whereas 3.28% occurred among populations. No significant genealogical branches or clusters were recognized on the neighbor-joining tree. Intra-group variation among populations was significant (φ st=0.032 85, P<0.01). These results suggest that E. rhadinum populations in the East and South China Seas have developed divergent genetic structures. Tests of neutral evolution and mismatch distribution suggest that E. rhadinum may have experienced a population expansion. The present study provides basic information for the conservation and sustainable exploitation of this species.
Similar content being viewed by others
References
An H S, Jee Y J, Min K S, Kim B L, Han S J. 2005. Phylogenetic analysis of six species of pacific abalone (Haliotidae) based on DNA sequences of 16S rRNA and cytochrome c oxidase subunitImitochondrial genes. Marine Biotechnology, 7: 373–380.
Avise J C. 2004. Molecular Markers, Natural History, and Evolution, Seconded. Sinauer Associates, Sunderland, (Massachusetts). p.686.
Baldwin B S, Black M, Sanjur O, Gustafson R, Lutz R A, Vrijenhoek R C. 1996. A diagnostic molecular marker for zebra mussels (Dreissena polymorpha) and potentially co-occurring bivalves: mitochondrial COI. Molecular Marine Biology and Biotechnology, 5: 9–10.
Chang N N, Shiao J C, Gong J C. 2012. Diversity of demersal fish in the East China Sea: implication of eutrophication and fishery. Continental Shelf Research, 47: 42–54.
Excoffier L, Laval G, Schneider S. 2008. Arlequin ver 3.1: An Integrated Software Package for Population Genetics Data Analysis. University of Bern, Bern, Switzerland.
Fu Y X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147: 915–925.
Grant W, Bowen B. 1998. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89: 415–426.
Grant W S, Spies I B, Canino M F. 2006. Biogeographic evidence for selection on mitochondrial DNA in North Pacific Walleye Pollock Theragra chalcogramma. Journal of Heredity, 97: 571–580.
Gruenthal K M, Acheson L K, Burton R S. 2007. Genetic structure of natural populations of California red abalone (Haliotis rufescens) using multiple genetic markers. Marine Biology, 152: 1 237–1 248.
Harpending H. 1994. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology, 66: 591.
Habib K A, Jeong D, Myoung J G, Kim M S, Jang Y S, Shim J S, Lee Y H. 2011. Population genetic structure and demographic history of the fat greenling Hexagrammos otakii. Genes & Genomics, 33: 413–423.
Haney R A, Silliman B R, Rand D M. 2010. Effects of selection and mutation on mitochondrial variation and inferences of historical population expansion in a Caribbean reef fish. Molecular Phylogenetics and Evolution, 57: 821–828.
Harpending H C, Sherry S T, Rogers A R, Stoneking M. 1993. The genetic structure of ancient human populations. Current Anthropology, 34: 483–496.
Han Z Q, Gao T X, Yanagimoto T, Sankurai Y. 2008. Genetic population structure of Nibea albiflora in Yellow Sea and East China Sea. Fisheries Science, 74: 544–552.
Huang G Y, Zhang T, Zhao F, Huang R, Yang Y, Zhang L Z. 2012. On morphology and histology of the digestive tract in young Eleutheronema rhadinum. Marine Fisheries, 34: 154–162. (in Chinese with English abstract)
Horne J B, Momigliano P, Welch D J, Newman S J, Herwerden L V. 2011. Limited ecological population connectivity suggests low demands on self-recruitment in a tropical in shore marine fish (Eleutheronema tetradactylum: Polynemidae). Molecular Ecology, 20: 2 291–2 306.
Kim W J, Kim K K, Han H S, Nam B H, Kim Y O, Kong H J, Noh J K, Yoon M. 2010. Population structure of the olive flounder (Paralichthys olivaceus) in Korea inferred from microsatellite marker analysis. Journal of Fish Biology, 76: 1 958–1 971.
Liu J X, Gao T X, Wu S F, Zhang Y P. 2007. Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus. (Temminck & Schlegel, 1845). Molecular Ecology, 16: 275–288.
Lynch M, Crease T J. 1990. The analysis of population survey data on DNA sequence variation. Molecular Biology and Evolution, 7: 377–394.
Liu Z J, Cordes J. 2004. DNA marker technologies and their applications in aquaculture genetics. Aquaculture, 238: 1–37.
Liu F, Xia J H, Bai Z Y, Fu J J, Li J L, Yue G H. 2009. High genetic diversity and substantial population differentiation in grass carp (Ctenopharyngodon idella) revealed by microsatellite analysis. Aquaculture, 297: 51–56.
Machado-Schiaffino G, Garcia-Vazquez E. 2011. Population structure of long tailed hake Macruronus magellanicus in the Pacific and Atlantic oceans: implications for fisheries management. Fisheries Research, 111: 164–169.
Ma C Y, Cheng Q Q, Zhang Q Y, Zhuang P, Zhao Y L. 2010. Genetic variation of Coilia ectenes (Clupeiformes: Engraulidae) revealed by the complete cytochrome b sequences of mitochondrial DNA. Journal of Experimental Marine Biology and Ecology, 385: 14–19.
Moore B R, Stapley J, Allsop Q, Newman S J, Ballagh A, Welch D J, Lester R J G. 2011. Stock structure of blue threadfin Eleutheronema tetradactylum across northern Australia, as indicated by parasites. Journal of Fish Biology, 78: 923–936.
Motomura H, Iwatsuki Y, Kimura S, Yoshino T. 2002. Revision of the Indo-West Pacific polynemid fish genus Eleutheronema (Teleostei: Perciformes). Ichthyological Research, 49: 47–61.
Nei M. 1987. Molecular Evolutionary Genetics. Columbia University Press, New York, USA. p.10-88.
Peng S M, Shi Z H, Hou J L, Wang W, Zhao F, Zhang H. 2009. Genetic diversity of silver pomfret (Pampus argenteus) populations from the China Sea based on mitochondrial DNA control region sequences. Biochemical Systematics and Ecology, 37: 626–632.
Röhl A, Mihn D. 2003. Network: A Program Package for Calculating Phylogenetic Networks. Mathematisches Seminar, University of Hamburg, Hamburg.
Rogers A R, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution, 9: 552–569.
Rogers A R. 1995. Genetic evidence for a Pleistocene population explosion. Evolution, 49: 608–615.
Rozas J, Sánchez-DelBarrio J C, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19: 2 496–2 497.
Spies I B, Gaichas S, Stevenson D E, Orr J W, Canino M F. 2006. DNA-based identification of Alaska skates (Amblyraja, Bathyraja and Raja: Rajidae) using cytochrome c oxidase subunit I (COI) variation. Journal of Fish Biology, 69: 283–292.
Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4: 406–425.
Schneider S, Excoffier L. 1999. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics, 152: 1 079–1 089.
Sun P, Shi Z H, Yin F, Peng S M. 2012. Genetic variation analysis of Mugil cephalus in China Sea based on mitochondrial COI gene sequences. Biochemical Genetics, 50: 180–191.
Shui B N, Han Z Q, Gao T X, Miao Z Q, Yanagimoto T. 2009. Mitochondrial DNA variation in the East China Sea and Yellow Sea populations of Japanese Spanish mackerel Scomberomorus niphonius. Fisheries Science, 75: 593–600.
Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123: 585–595.
Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24: 1 596–1 599.
Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25: 4 876–4 882.
Ward R D, Zemlak T S, Innes B H, Last P R, Hebert P D N. 2005. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society B: Biological Sciences, 360: 1 847–1 857.
Xiao Y S, Zhang Y, Gao T X, Yanagimo T, Yabe M, Sakurai Y. 2009. Genetic diversity in the mtDNA control region and population structure in the small yellow croaker Larimichthys polyactis. Fisheries Science, 85: 303–314.
Xu D D, Lou B, Shi H L, Geng Z, Li S L, Zhang Y R. 2012. Genetic diversity and population structure of Nibea albiflora in the China Sea revealed by mitochondrial COI sequences. Biochemical Systematics and Ecology, 45: 158–165.
You F, Wang K, Xiang J, Xu C. 2001. Comparative analysis of biochemical genetic structure and variance between natural and cultured stocks on the left-eyed flounder, Paralichthys olivaceus off Shandong coastal waters. Oceanologia et Limnologia Sinica, 32: 512–512. (in Chinese with English abstract)
Yue G H, Zhu Z Y, Lo L C, Wang C M, Lin G, Feng F, Pang H Y, Li J, Gong P, Liu H M. 2009. Genetic variation and population structure of Asian seabass (Lates calcarifer) in the Asia-Pacific region. Aquaculture, 293: 22–28.
Zischke M T, Cribb T H, Welch D J, Sawynok W, Lester R J G. 2009. Stock structure of blue threadfin Eleutheronema tetradactylum on the Queensland east coast, as determined by parasites and conventional tagging. Journal of Fish Biology, 75: 156–171.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supported by the National Key Technology Research and Development Program of China (No. 2011BAD13B08) and the Central Nonprofit Basic Scientific Research Project for the Scientific Research Institutes of China (No. 2011Z01)
Rights and permissions
About this article
Cite this article
Sun, X., Xu, D., Lou, B. et al. Genetic diversity and population structure of Eleutheronema rhadinum in the East and South China Seas revealed in mitochondrial COI sequences. Chin. J. Ocean. Limnol. 31, 1276–1283 (2013). https://doi.org/10.1007/s00343-013-3005-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00343-013-3005-2