Abstract
Although Northern red oak (Quercus rubra L.) is the most important introduced deciduous tree species in Germany, only little is known about its genetic variation. For the first time, we describe patterns of neutral and potentially adaptive nuclear genetic variation in Northern red oak stands across Germany. For this purpose, 792 trees were genotyped including 611 trees from 12 stands in Germany of unknown origin and 181 trees from four populations within the natural distribution area in North America. Our marker set included 12 potentially adaptive (expressed sequence tag-derived simple sequence repeat = EST SSR) and 8 putatively selectively neutral nuclear microsatellite (nSSR) markers. Our results showed that German stands retain comparatively high levels of genetic variation at both EST-SSRs and nSSRs, but are more similar to each other than to North American populations. These findings are in agreement with earlier chloroplast DNA analyses which suggested that German populations originated from a limited geographic area in North America. The comparison between potentially adaptive and neutral microsatellite markers did not reveal differences in the analyzed diversity and differentiation measures for most markers. However, locus FIR013 was identified as a potential outlier locus. Due to the absence of signatures of selection in German stands, we suggest that introduced populations were established with material from provenances that were adapted to environmental conditions similar to those in Germany. However, we analyzed only a limited number of loci which are unlikely to be representative of adaptive genetic differences among German stands. Our results suggest that the apparent introduction from a limited geographic range in North America may go along with a reduced adaptive potential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The introduction of Northern red oak (Quercus rubra L.) from its natural range in North America to Europe dates back to the end of the seventeenth century, when it was likely first brought to France (Houba 1887; Hickel 1932). Until the middle of the eighteenth century, Q. rubra was planted in parks and botanical gardens for ornamental purposes (Bauer 1951; Nagel 2015). After two major waves of cultivation in the second half of the nineteenth century and at the turn of the nineteenth to the twentieth century, it is now the most important non-native deciduous tree species for wood production in Germany (Bauer 1951; Bundesministerium für Ernährung und Landwirtschaft (BMEL) 2014). Nagel (2015) described Q. rubra as a species covering a wide range of soil and climatic conditions with annual precipitation between 600 and 2000 mm and mean annual temperatures between 4 and 15 °C and featuring a shorter rotation period (~ 80–120 years) than native white oak species (~ 140 years), as well as a lower demand for nutrients and water.
Unlike other European countries (Möllerová 2005; Riepšas and Straigytė 2008; Chmura 2013), Q. rubra is not considered invasive in Germany. It is less shade-tolerant than the main native tree species Fagus sylvatica L. and only little more shade tolerant than the native white oak species Q. robur L. (Vor and Lüpke 2004; Niinemets and Valladares 2006; Nagel 2015). Moreover, it is easily controllable by containment measures (Vor 2005; Nagel 2015).
Introduced species are usually expected to experience a founder effect and genetic bottlenecks that greatly promote genetic drift and can result in decreased genetic variation, especially if only a limited numbers of trees served as seed source and/or only a small part of the species’ natural range was sampled (Nei et al. 1975; Barrett and Husband 1990). In the past, several studies were conducted to reveal patterns of genetic variation within Northern red oak’s natural range (e.g., Daubree and Kremer 1993; Romero-Severson et al. 2003; Magni et al. 2005; Zhang et al. 2015; Borkowski et al. 2017). However, only few studies focussed on the impact of the introduction on genetic variation of Northern red oak populations in Europe (Magni Diaz 2004; Merceron et al. 2017; Pettenkofer et al. 2019).
Borkowski et al. (2017) found that the genetic differentiation between Northern red oak populations increased from south to north within the natural range, reflecting its postglacial migration movement, but without revealing distinct pathways. Magni Diaz (2004) used chloroplast (cp) DNA-based PCR–RFLP (restriction fragment length polymorphism) markers to analyze the genetic variation of introduced stands mainly located in France and Germany with only a few samples from the Netherlands, Belgium, Spain, Italy, and Romania. Interestingly, Magni Diaz (2004) found greater total and within-population diversity as well as differentiation between populations for historical German populations in comparison with historical French populations potentially as a result of different import and forest policies in these countries. However, for all European populations, Magni Diaz (2004) could not identify a geographic pattern. Recently, Pettenkofer et al. (2019) used maternally inherited chloroplast DNA markers to analyze the genetic variation of German stands in comparison with North American populations. Most German stands showed very similar haplotype frequencies and low haplotype diversity, pointing to a limited number of seed sources. A considerably higher cpDNA haplotype diversity was detected in Southern Germany. Multiple introductions and admixture of material within Europe or prior to its introduction to Europe were suggested as possible reasons for high haplotype diversity within certain regions (Magni Diaz 2004; Pettenkofer et al. 2019). While it was not possible to narrow down the geographic origin to specific regions in North America, patterns of genetic variation suggested an origin from a limited geographic area, likely in the northern part of the species’ distribution range (Magni Diaz 2004; Pettenkofer et al. 2019).
Merceron et al. (2017) studied single-nucleotide polymorphism (SNP) markers randomly distributed across the genome in European populations, mainly in France, but also including samples from Germany, the Netherlands, Belgium, Spain, Italy, and Romania, and compared them with data for North American populations. Three main genetic clusters were identified in North America: in the south, the northeast, and the northwest, respectively. However, only trees representing the two northern clusters were found in Europe. Merceron et al. (2017) stated that trees representing the southern cluster were either never introduced to Europe or vanished eventually. They suggested that European populations may thus originate from the northern part of the natural range, a conclusion supported by other studies on this topic (Bauer 1954; Magni Diaz 2004; Nagel 2015; Pettenkofer et al. 2019).
This study used some of the same populations that were analyzed earlier with chloroplast SSR markers (Pettenkofer et al. 2019). In contrast to chloroplast SSR markers, which are maternally inherited in Q. rubra and therefore useful to track migration routes (Petit et al. 1997; Alexander and Woeste 2014), nSSR markers are biparentally inherited and much more polymorphic. However, nSSR markers require a considerably larger set of samples per population. For the first time, we describe patterns of neutral and potentially adaptive nuclear genetic variation in Northern red oak stands across Germany.
The main objective of the present study was to assess the genetic variation of German Northern red oak stands by analyzing variable potentially adaptive (expressed sequence tag-derived simple sequence repeats = EST SSR) and putatively selectively neutral nuclear microsatellite (nSSR) markers and to compare German stands with selected North American populations.
We hypothesize that (1) the Q. rubra gene pool introduced to Germany represents only a fraction of the North American gene pool, and (2) potentially adaptive genetic markers show different variation patterns compared to neutral markers.
Materials and methods
Plant material
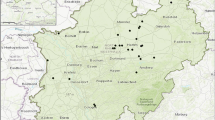
In total, 792 trees were genotyped in this study including 611 trees from 12 stands in Germany of unknown origin (48–57 samples per stand; Table 1, Fig. 1) and 181 trees from four populations within the natural distribution area in North America (40–47 samples per population; Table 2, Fig. 2). The selection of the study sites was based on the chloroplast haplotype frequencies identified earlier in these populations based on smaller sample sizes (5–10 samples per population) (Pettenkofer et al. 2019). Due to the strict maternal inheritance and lower variation of cpDNA, these sample sizes allowed to characterize 39 stands in Germany and 19 populations in the USA and Canada. In the study carried out by Pettenkofer et al. (2019), most German stands had the most common haplotype “A.” The next frequent haplotype “B” was found exclusively in the northern parts of the natural range. Two of the three haplotype clusters identified in the natural range were also found in German populations. Stands located in the southwest of Germany had a higher haplotype variation compared to other sampled stands. The populations within the natural range used in this study were selected from the northern and southern parts of the range that were different based on the chloroplast haplotypes, and samples were available in sufficient numbers for them. Samples were taken randomly in each population as buds, green leaves, or cambium probes.
 )
)Within Germany, sample stands were selected in five different federal states of Germany (Table 1): two stands in each of Lower Saxony, North Rhine-Westphalia, Brandenburg, and Thuringia and four stands in Baden-Wuerttemberg, where Pettenkofer et al. (2019) discovered the highest chloroplast variation. All German stands were (1) pure Q. rubra stands, (2) 50–80 years old, (3) featured a rectangular shape for easier data acquisition, and (4) both the present and expected future climate conditions matched their autecological properties.
DNA isolation
The DNeasy 96 Plant Kit (Qiagen, Hilden, Germany) was used to extract the DNA from either about 1 cm2 leaf tissue of the fresh leaf or 1–2 whole buds from a fresh twig per tree.
Extraction of DNA from cambium samples was performed with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) using the respective manufacturer’s protocol with the following changes: The incubation of cambium samples took place overnight at 65 °C in a lysate containing the buffer AP1 and 9.4% polyvinylpyrrolidone (PVP) in the final solution.
EST- and neutral nuclear SSR markers
For initial testing, we selected 25 nuclear microsatellite markers arranged in 4 multiplexes (Supplementary Table 1S). Five of them (FIR043, FIR004, quru-GA-2M04, quru-GA-2F05, and GOT037) were removed due to difficulties in amplification or allele scoring. Finally, we used 12 EST-SSR and 8 neutral nSSR markers (Aldrich et al. 2002; Durand et al. 2010; Sullivan et al. 2013; Supplementary Table 1S).
Before the preparation of the specific multiplexes, each forward and reverse primer was diluted to a concentration of 5 pM/µl. Except for multiplex 4 (as explained further below), the PCR mix contained for each sample the following components: 1 µl of genomic DNA (about 0.6 ng/µl), 1.5 µl PCR buffer (containing 0.8 M Tris–HCl and 0.2 M (NH4)2SO4), 1.5 µl MgCl2 (25 mM), 1 µl of each dNTP (2.5 mM), and 1 U HOT FIREPol®Taq-polymerase (Solis BioDyne; Tartu, Estonia). In addition, the following components were added to their specific multiplex in the denoted amount: multiplex 1–0.8 µl ddH2O and 1 µl of each forward and reverse primer for the markers quru-GA-1P10, quru-CA-2P24, quru-CA-3A05, and quru-CA-3D15, respectively; multiplex 2–0.8 µl ddH2O and 1 µl of each forward and reverse primer for the markers FIR043, FIR035, FIR028, and FIR013, respectively; multiplex 3—two separate PCRs were performed—(1) 4.8 µl ddH2O and the denoted amount for each forward and reverse primer for the markers quru-GA-0E09 (1 µl), quru-GA-C06 (0.6 µl), and quru-GA-F07 (0.4 µl), (2), 3.8 µl ddH2O and the denoted amount for each forward and reverse primer for the markers GOT021 (0.8 µl), FIR024 (0.8 µl), FIR031 (0.5 µl), and quru-GA-2M04 (0.4 µl). The products of the last two PCRs were pooled before electrophoretic separation.
Multiplex 4 was performed with the following PCR mix per sample: 8.5 µl Multiplex PCR Kit (Qiagen, Hilden, Germany) and the denoted amount for each forward and reverse primer for the markers PIE040 (1 µl), GOT037 (1 µl), quru-GA-2F05 (0.6 µl), quru-GA-0C11 (0.4 µl), FIR104 (0.4 µl), PIE125 (0.4 µl), VIT107 (0.4 µl), VIT023 (0.4 µl), and GOT040 (0.3 µl).
The same touchdown PCR program was used for all markers in a Biometra TProfessional thermocycler (Jena, Germany). The PCR protocol started with 15 min for initial denaturation at 95 °C, followed by 10 cycles of 1-min denaturation at 94 °C, 1-min annealing at 60 °C (− 1 °C per cycle), and 1-min extension at 72 °C. This first set of cycles was then followed by another 25 cycles of 1-min denaturation at 94 °C, 1-min annealing at 50 °C, and 1-min extension at 72 °C. The PCR ended with a final 20-min extension step.
Before adding to HiDi formamide, the PCR products were diluted for capillary gel electrophoresis based on band intensities on agarose gels in the ratios of 1:150 for multiplexes 1 and 2 and 1:60 for multiplexes 3 and 4. The SSR fragments were separated using capillary electrophoresis on an ABI Genetic Analyzer 3130xl (Applied Biosystems, Foster City, USA) and sized using the internal size standard GeneScan™ 500 ROX™ as reference for multiplexes 1 and 2 and GeneScan™ 500 LIZ™ as reference for multiplexes 3 and 4 (Applied Biosystems, Foster City, USA). The fragments were scored using the software package GeneMapper version 3.7 (Applied Biosystems, Foster City, USA).
Data analyses
Based on the obtained SSR genotypes, the software GenAlEx 6.5 (Peakall and Smouse 2006, 2012) was used to calculate the number of alleles Na, the effective number of alleles Ne, the observed heterozygosity Ho, the expected heterozygosity He, and the genetic differentiation measures D (Jost 2008) and FST for all markers and populations. We chose to calculate both differentiation parameters because traditional FST can underestimate differentiation for such highly polymorphic markers as SSRs (Jost 2008). The fixation index FIS and the test for significant differences of its values from 0 were calculated with FSTAT 2.9.3 with P-values adjusted using the sequential Bonferroni procedure (Rice 1989; Goudet 1995). The Kruskal–Wallis test was used to identify significant differences between German and North American populations for all diversity parameters (Table 3) using the software R version 3.3.2 (R Core Team 2016). Also, the Kruskal–Wallis test with multiple comparisons implemented in the R-package “pgirmess” (Giraudoux et al. 2018) was used to test for pairwise differences among all populations. The implemented multiple-comparison test determines which population pair shows significant differences. To visualize population differences, we further used GenAlEx 6.5 to perform a principal coordinates analysis (PCoA). This cluster analysis was based on the pairwise genetic distance matrix between populations and between individuals. The PCoA assigns a location for each individual or population within a multidimensional space. Two or three first main axes that explain most of the genetic differentiation between individuals and/or populations are usually presented in a plot based on this analysis (Peakall and Smouse 2012). In this study, the genetic distance matrix was based on Jost’s D (Jost 2008) calculated using GenAlEx 6.5.
To build a neighbor-joining tree (NJT), the genetic distance DA by Nei et al. (1983), which is especially suited for microsatellite markers (Takezaki and Nei 1996), was calculated using the Populations 1.2.32 software (Langella 1999). The bootstrap values were based on 1000 permutations across loci, and the NJT was visualized using the online software IcyTree (Vaughan 2017).
Arlequin version 3.5 (Excoffier and Lischer 2010) was also used to perform an analysis of molecular variance (AMOVA) by applying 9999 permutations for all populations (the entire dataset), all German populations, and the four North American populations (Excoffier et al. 1992). The software computes a matrix of pairwise distances between all populations using the number of different alleles (FST-like). Further, Arlequin was also used to search for outlier loci that could be potentially under selection (Excoffier et al. 2009). Here, 50,000 simulations and 100 demes were selected as running conditions.
To make inferences about the potential population structure and number of clusters that can be identified in our genotyped samples, we used the software STRUCTURE 2.3.4 by Pritchard et al. (2000). STRUCTURE uses a model-based statistical clustering method within a Bayesian framework to assign individuals to populations and detect population structure over all sampled multilocus genotypes (Pritchard et al. 2000; Falush et al. 2003). We used 10,000 and 100,000 Markov chain Monte Carlo (MCMC) replicates for the burn-in period and further iterations, respectively. Runs were performed for 1–20 potential clusters (K) using 20 iterations for each test. To infer the optimal number of K, we applied the ΔK method by Evanno et al. (2005) using the STRUCTURE HARVESTER 0.6.94 software (Earl and Von Holdt 2012). Results were visualized using the online software CLUMPAK (Kopelman et al. 2015).
To test for genetic bottleneck effects, the software BOTTLENECK (Cornuet and Luikart 1996) was used with 1000 iterations and assuming a two-phase model (TPM). The TPM allows to assume different proportions (contributions) of the infinite allele model (IAM) and the stepwise mutation model (SMM) for all markers. Considering that dinucleotide and other perfect repeats follow the IAM while imperfect repeats rather follow the SMM, we selected the option with 70% IAM and 30% SMM in accordance with our marker set (Cornuet and Luikart 1996; Cristescu et al. 2010).
Results
Genetic diversity
The German stands and North American populations showed similar levels of mean observed (Ho = 0.61 vs. 0.59) and expected (He = 0.63 vs. 0.65) heterozygosities, respectively (Table 3). These parameters were also similar across all populations (Ho = 0.57–0.62, He = 0.62–0.68, Table 3). The mean (Na) and effective (Ne) number of alleles also showed no significant differences between German and North American populations (Table 3). However, the number of private alleles and their mean frequency in Germany (9 in 12 populations, Np = 0.75) were significantly lower than those in the North American populations (19 in 4 populations, Np = 4.75) (Tables 3 and 4). In Germany, 5 of 9 private alleles were found in stands in Baden-Wuerttemberg, while two were found in each Brandenburg and Lower Saxony (Table 4). We detected no private alleles in stands from Thuringia or North Rhine-Westphalia. In North America, most private alleles were found in BR1 (Np = 9) and Constance Bay (Np = 5); both populations are located in the north of the natural distribution range. Relative frequencies of private alleles range from 0.009 to 0.042 (Table 4). Among all populations and tested parameters (Na, Ne, Ho, He, FIS), only FIS showed significant differences between the populations 18 (North Rhine-Westphalia) and BR1. No recent genetic bottleneck was detected using the BOTTLENECK software.
The search for outlier loci that could be potentially under selection identified only the EST-SSR FIR013 as a significant outlier locus (Supplementary Table 2S and Fig. 3). At this marker, the North American population BR1 shows a higher frequency (0.24) of allele 138 in comparison with other populations (0.01–0.11).
Genetic differentiation and population structure
The PCoA based on Jost’s D between all populations showed that German stands densely clustered between the North American population FC-B, which is located in the northwest of the natural range (Ford Forestry Center, Michigan), and Nantahala, which is located in the south of the range, in the western parts of North Carolina (Fig. 4). Constance Bay (Canada) and BR1 (Brockway Mountain, Michigan) were set apart from the German cluster. The first two principal coordinates together explained 66.32% of the variation among populations. Also, the individual-based PCoA revealed more heterogeneous point clouds for the populations of North American origin as compared to the German plantations. Especially, the two natural populations from Northern Michigan occupy a comparatively wide space in the two-dimensional PCoA (Supplementary figures 5S and 6S).
To partition genetic variation among and within populations, a hierarchical AMOVA was performed for all populations (the entire dataset) and separately for the North American populations and all German populations (Supplementary Tables 3S, 4S, and 5S). In the entire dataset, 1.10% of the variation was due to partition among populations, while 98.90% was due to variation within populations (among and within individuals), with all components being highly significant. In North America, 1.88% of the variation was due to partition among populations (p < 0.001), and 98.12% of the variation was due to variation within populations. In German populations, the genetic variation among populations was lower than among North American populations (0.78%).
The STRUCTURE and Delta K analyses of the entire dataset, as well as for North American and German populations, indicated no strong population structure (Supplementary figures 1S–4S).
The NJT did not reveal strong differentiation among German populations (Fig. 5) and was in agreement with the STRUCTURE and PCoA analyses. Most clusters were not well supported by bootstrap values. However, all four North American populations, BR1, FC-B, Nantahala, and Constance Bay clustered together. The German population 31 clustered close to Nantahala and Constance Bay populations, but this relationship was not well supported.
Neighbor-joining tree (NJT) of the analyzed populations based on the genetic distance DA by Nei et al. (1983). Only bootstrap numbers above 50% are shown
Discussion
Genetic variation at EST-SSRs and nSSRs
Unlike nSSR markers, EST-SSRs are more conserved being located very close to functional genes whose variation is very likely to be limited due to selection. Thus, they feature higher transferability across species within genera than nSSR markers, but are not as polymorphic as nSSR markers (Ellis and Burke 2007; Kalia et al. 2011). Accordingly, for our dataset, mean allelic richness was lower for EST-SSR markers (Na = 5.63) than for the neutral nSSR markers (Na = 15.04). This pattern was also observed for the observed and expected heterozygosity (neutral: Ho = 0.798, He = 0.850; EST: Ho = 0.474, He = 0.494). Although we identified EST-SSR marker FIR013 as an outlier locus, in general, FST and Jost’s D were similar for potentially adaptive EST-SSR and neutral nSSR markers.
Genetic variation patterns indicate the origin of German populations from a limited area
Our results show that 19 rare alleles (relative frequencies = 0.010–0.042), which are private in the examined North American populations, are not found in German populations. These drift-like effects can be attributed to the selection and sampling of seed sources. The occurrence of other rare alleles in German stands and the uniform values of Ho and He across populations show no signs of a recent reduction of the overall genetic variation in German stands. Moreover, Ho and He remained uniform across all populations showing no signs of recent bottleneck effects. The lack of indications for recent bottleneck effects does not exclude that European Northern red oak populations might have undergone bottleneck effects in Germany at all. A genetic bottleneck could have been induced a long time ago, for instance due to selective sampling within the native range. However, admixture of material within Europe and multiple introductions over time could have maintained the high genetic variation in German populations (Magni Diaz 2004) but could also have led to a lower differentiation between introduced populations. Concerning the overall level of genetic variation in German Northern red oak stands, additional import of seeding material seems not to be necessary at the moment. But concerning the limited number of North American populations investigated in our study and with respect to future climate change scenarios, additional import of material from other parts of the natural range might be necessary in the future.
Interestingly, the number of private alleles among German populations was especially high in Baden-Württemberg, suggesting multiple introductions or admixture of plant material before its introduction (Pettenkofer et al. 2019). In North America, 14 out of 19 private alleles were found in the northern populations BR-1 (9) and Constance Bay (5).
The AMOVA showed that only 1.10% of the genetic variation was due to partitioning among populations. The PCoA clusters that visualized the genetic differentiation between populations (Fig. 4) and the pairwise population genetic distances supported the data of Pettenkofer et al. (2019). German populations clustered densely occupying only a small part of the two-dimensional space in the PCoA plot indicating the introduction from a limited geographic range (Fig. 4). Likewise, the distribution of point clouds in individual-based PCoA analyses and earlier chloroplast DNA analyses suggest that introduced populations originated from a limited range in the northern part of the species’ distribution.
Considering also the similar levels of genetic diversity in native and introduced populations and the absence of recent genetic bottlenecks, the clustering of populations in the PCoA supported the hypothesis that German stands might have been established with material from a particular region within the species’ native range.
Weak genetic structure was observed across all sampled populations
Although the bootstrap support was weak for most of the clusters, the NJT showed that the two North American populations that are located close to the Lake Superior in Michigan, USA (FC-B and BR1), are clustered together. For the North American cluster, however, bootstrap support was rather low (= 28%), showing only weak distinction between North American and German populations.
However, there is no strong evidence for inter-populational or inter-regional differentiation within both German stands and North American populations.
In this study, only the EST-SSR marker FIR013 was identified as potentially under selection. It was the most differentiated marker with the highest FST value among all markers (Supplementary Table 2S), mostly due to the different frequencies of the FIR013 alleles in the North American population BR1 (data not shown), which was also the most differentiated population among all populations with the highest pairwise Jost’s D and FST values. FIR013 is located in a CONSTANS-like gene and represents a region consisting of several tandemly repeated codons CAG and/or CAA encoding glutamine. This gene is known to be involved in the regulation of flowering time and photoperiodic control of growth (Lind-Riehl et al. 2014). Also, the distribution of alleles between populations of closely related Q. rubra and Q. ellipsoidalis shows that FIR013 might also serve as a marker to trace potential introgression of genes from Q. ellipsoidalis into Q. rubra (Lind-Riehl et al. 2014). Considering the overlap of ranges of both species in the Great Lakes region, genetic introgression may be one possible reason for higher differentiation of BR1. However, population BR1 is located outside the current distribution range of Q. ellipsoidalis (Lind-Riehl et al. 2014). The absence of signatures of local adaptation in German populations may suggest that introduced populations were established with material from provenances with similar environmental conditions as the ones prevailing in Germany (Liesebach and Schneck 2011; Pettenkofer et al. 2019). However, we only analyzed a limited number of potentially adaptive markers. Further analyses of candidate genes for local adaptation and of adaptive traits in provenance trials are needed to assess adaptive differences among provenances.
Conclusions and future perspective
Comparatively high levels of genetic variation at both EST-SSRs and nSSRs in German stands suggest that additional import of seeding material from the native range is not required for maintaining a sufficiently high level of genetic variation in Germany. Estimation of chloroplast haplotype diversity (Pettenkofer et al. 2019) further revealed that plantations in the southwest of Germany may provide a variable genetic resource. Moreover, Liesebach and Schneck (2011) state that German provenances perform better in respect of growth than provenances from the natural range. Therefore, additional import of material from the native range is not necessary to enhance the genetic variation or growth of local Northern red oak stands. The analysis of additional candidate genes for local adaptation and of adaptive traits (e.g., drought tolerance) in provenance trials is needed to assess adaptive differences among German and North American provenances. The identification and genotyping of large numbers of SNPs can be achieved by genotyping by sequencing techniques such as restriction site-associated DNA sequencing (RADseq) and thus provide a new source of genome-wide potentially adaptive markers (Miller et al. 2007; Davey and Blaxter 2010).
Finally, North American populations representing the natural range could be characterized at nuclear and chloroplast DNA markers to narrow down the geographic origin of German plantations.
References
Aldrich P, Michler C, Sun W et al (2002) Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol Ecol Notes 2:472–474
Alexander LW, Woeste KE (2014) Pyrosequencing of the northern red oak (Quercus rubra L.) chloroplast genome reveals high quality polymorphisms for population management. Tree Genet Genomes 10:803–812
Barrett SC, Husband BC (1990) The genetics of plant migration and colonization. In: Brown HD, Clegg MT, Kahler AL et al (eds) Plant population genetics, breeding, and genetic resources. Sinauer Associates Inc., Sunderland, pp 254–277
Bauer F (1951) Die Roteiche 1950. Eine Ertragskundlich-Biologische Und Holzkundliche Untersuchung. Dissertation, Universität Göttingen
Bauer F (1954) Zur Rassenfrage Der Roteiche. Allgemeine Forstzeitschrift 9:470–474
Borkowski DS, Hoban SM, Chatwin W et al (2017) Rangewide population differentiation and population substructure in Quercus rubra L. Tree Genet Genomes 13:472
Bundesministerium Für Ernährung Und Landwirtschaft (Bmel) (2014) Der Wald In Deutschland. Ausgewählte Ergebnisse Der Dritten Bundeswaldinventur. Available Via https://www.Bundeswaldinventur.De/Fileadmin/Site_Master/Content/Dokumente/Downloads/Bmel_Wald_Broschuere.Pdf. Accessed 20 Mar 2018
Chmura D (2013) Impact of alien tree species Quercus rubra L. on understorey environment and flora: a study of the Silesian Upland (southern Poland). Pol J Ecol 61:431–442
Cornuet J-M, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cristescu R, Sherwin WB, Handasyde K et al (2010) Detecting bottlenecks using BOTTLENECK 1.2.02 in wild populations. the importance of the microsatellite structure. Conserv Genet 11:1043–1049
Daubree jb, Kremer A (1993) Genetic and phenological differentiation between introduced and natural populations of Quercus rubra L. Ann For Sci 50:271s–280s
Davey JW, Blaxter M (2010) Radseq. next-generation population genetics. Brief Funct Genom 9:416–423
Durand J, Bodénès C, Chancerel E et al (2010) A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genom 11:570
Earl DA, Von Holdt BM (2012) STRUCTURE HARVESTER. A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellis JR, Burke JM (2007) EST-SSRS as a resource for population genetic analyses. Heredity 99:125–132
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure. A simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HLE (2010) Arlequin suite ver 3.5. A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro M (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 2:479–491
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity 103:285–298
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Giraudoux P, Antonietti J-P, Beale C et al (2018) Pgirmess (R-package). Spatial analysis and data mining for field ecologists. https://cran.r-project.org/web/packages/pgirmess/index.html
Goudet J (1995) Fstat (Version 1.2). A computer program to calculate F-statistics. J Heredity 86:485–486
Hickel R (1932) Dendrologie forestière. Lechevalier, Paris
Houba J (1887) Les chênes de l’amérique septentrionale en Belgique. leur origine, leur qualités, leur avenir. In: Ceysens M. (ed) Hasselt
Jost L (2008) Gst and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kalia RK, Rai MK, Kalia S et al (2011) Microsatellite markers. An overview of the recent progress in plants. Euphytica 177:309–334
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak. A program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Langella O (1999) Populations. Cnrs Upr 9034, Gif-Sur-Yvette Cedex
Liesebach M, Schneck V (2011) Entwicklung Von Amerikanischen Und Europäischen Herkünften Der Roteiche in Deutschland. Forstarchiv 82:125–133
Lind JF, Gailing O (2013) Genetic structure of Quercus rubra L. and Quercus ellipsoidalis E. J. Hill populations at gene-based EST-SSR and nuclear SSR markers. Tree Genet Genomes 9:707–722
Lind-Riehl JF, Sullivan AR, Gailing O (2014) Evidence for selection on a CONSTANS-like gene between two red oak species. Ann Bot 113:967–975
Magni CR, Ducousso A, Caron H et al (2005) Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Mol Ecol 14:513–524
Magni Diaz CR (2004) Reconstitution de L’introduction de Quercus rubra L. En Europe et conséquences génétiques dans les populations allochtones. Dissertation, Ècole Nationale Du Génie Rural, Des Eaux Et Des Forêts (In French)
Merceron NR, Leroy T, Chancerel E et al (2017) Back to America. tracking the origin of European introduced populations of Quercus rubra L. Genome 60:778–790
Miller MR, Dunham JP, Amores A et al (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17:240–248
Möllerová J (2005) Notes on invasive and expansive trees and shrubs. J For Sci 51:19–23
Nagel R-V (2015) Roteiche (Quercus rubra L.). In: Vor T, Spellmann H, Bolte A et al (eds) Potenziale und Risiken eingeführter Baumarten. Baumartenportraits Mit Naturschtuzfachlicher Bewertung. Univ.-Verl. Göttingen, Göttingen, pp 219–267
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. Ii. Gene frequency data. J Mol Evol 19:153–170
Niinemets Ü, Valladares F (2006) Tolerance to shade, drought, and waterlogging of temperate Northern Hemisphere trees and shrubs. Ecol Monogr 76:521–547
Peakall R, Smouse PE (2006) Genalex 6. Genetic analysis in Excel. population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) Genalex 6.5. Genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539 (Oxford, England)
Petit RJ, Pineau E, Demesure B et al (1997) Chloroplast DNA footprints of postglacial recolonization by oaks. Proc Natl Acad Sci USA 94:9996–10001
Pettenkofer T, Burkardt K, Ammer C et al (2019) Genetic diversity and differentiation of introduced red oak (Quercus rubra) in Germany in comparison with reference native North American populations. Eur J For Res 38:275–285
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2016) R. A language and environment for statistical computing. R Foundation For Statistical Computing, Vienna
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Riepšas E, Straigytė L (2008) Invasiveness and ecological effects of red oak (Quercus rubra L.) in lithuanian forests. Baltic For 14:122–130
Romero-Severson J, Aldrich P, Feng Y et al (2003) Chloroplast DNA variation of northern red oak (Quercus rubra L.) in Indiana. New For 26:43–49
Sullivan AR, Lind JF, Mccleary TS et al (2013) Development and characterization of genomic and gene-based microsatellite markers in North American red oak species. Plant Mol Biol Rep 31:231–239
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Vaughan TG (2017) IcyTree. Rapid browser-based visualization for phylogenetic trees and networks. Bioinformatics 33:2392–2394 (Oxford, England)
Vor T (2005) Natural regeneration of Quercus rubra L. (red oak) in Germany. Neobiota 6:111–123
Vor T, Lüpke BV (2004) Das Wachstum von Roteiche, Traubeneiche und Rotbuche unter verschiedenen Lichtbedingungen in den ersten beiden Jahren nach der Pflanzung. Forstarchiv 75:13–19
Zhang R, Hipp AL, Gailing O (2015) Sharing of chloroplast haplotypes among red oak species suggests interspecific gene flow between neighboring populations. Botany 93:691–700
Acknowledgements
Open Access funding provided by Projekt DEAL. We thank Alexandra Dolynska, Andreas Parth, Katharina Burkardt, Christine Radler, and Natalie Breidenbach for their valuable technical assistance and Mirko Liesebach (Thünen Institute) for providing plant material. The study was supported by the German Federal Ministry of Food and Agriculture (Funding Code 22023314).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Rüdiger Grote.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pettenkofer, T., Finkeldey, R., Müller, M. et al. Genetic variation of introduced red oak (Quercus rubra) stands in Germany compared to North American populations. Eur J Forest Res 139, 321–331 (2020). https://doi.org/10.1007/s10342-019-01256-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10342-019-01256-5