Abstract
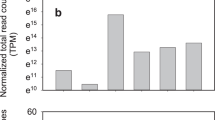
Multiple studies have attempted to dissect the molecular mechanism underlying seed development in chickpea (Cicer arietinum L.). These studies highlight the need to focus on the role of miRNAs in regulating storage protein accumulation in seeds. Therefore, a total of 8,856,691 short-read sequences were generated from a small RNA library of developing chickpea seeds and were analyzed using miRDeep-P to identify 74 known and 26 novel miRNA sequences. Known miRNAs were classified into 22 miRNA families with miRNA156 family being most abundant. Of the 26 putative novel miRNAs identified, only 22 could be experimentally validated using stem loop end point PCR. Differential expression analyses led to the identification of known as well as novel miRNAs that could regulate various stages of chickpea seed development. In silico target prediction revealed several important target genes and transcription factors like SPL, mediator of RNA Polymerase II transcription subunit 12, aspartic proteinase and NACs, which were further validated by real-time PCR analysis. A comparative expression analysis in chickpea genotypes with contrasting seed protein content revealed one known (Car-miR156h) and two novel miRNA (Car-novmiR7 and Car-novmiR23) candidates to be highly expressed in the LPC (low protein content) chickpea genotypes, targets of which are known to regulate seed storage protein accumulation. Therefore, this study provides a useful resource in the form of miRNA and their targets which can be further utilized to understand and manipulate various regulatory mechanisms involved in seed development with the overall aim of improving yield and nutrition attributes in chickpea.






Similar content being viewed by others
Data availability
All relevant supporting data can be found within the additional files accompanying this article. The RNA-Seq reads described in the article were deposited at the Sequence Read Archive (SRA) database; BioProject PRJNA298120; Accession number SRR3103025.
Abbreviations
- RPM:
-
reads per million
- TF:
-
transcription factor
- LPC:
-
low protein content
- HPC:
-
high protein content
- GO:
-
gene ontology
References
Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci 104(41):16371–16376
Bai B, Shi B, Hou N, Cao Y, Meng Y, Bian H, Zhu M, Han N (2017) microRNAs participate in gene expression regulation and phytohormone cross-talk in barley embryo during seed development and germination. BMC Plant Biol 17(1):150
Ballen-Taborda C, Plata G, Ayling S, Rodriguez-Zapata F, Becerra Lopez-Lavalle LA, Duitama J, Tohme J (2013) Identification of cassava MicroRNAs under abiotic stress. Int J Genomics 2013:857986–857910. https://doi.org/10.1155/2013/857986
Bao W, Kojima KK, Kohany O (2015) Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile Dna 6(1):11
Bellieny-Rabelo D, De Oliveira EA, da Silva RE, Costa EP, Oliveira AE, Venancio TM (2016) Transcriptome analysis uncovers key regulatory and metabolic aspects of soybean embryonic axes during germination. Sci Rep 6:36009
Berezikov E, Cuppen E, Plasterk RH (2006) Approaches to microRNA discovery. Nature Genet 38(6):S2–S7
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320(5880):1185–1190
Cao R, Guo L, Ma M, Zhang W, Liu X, Zhao H (2019) Identification and functional characterization of Squamosa promoter binding protein-like gene TaSPL16 in wheat (Triticum aestivum L.). Front Plant Sci 10(212)
Cao S, Zhu QH, Shen W, Jiao X, Zhao X, Wang MB, Liu L, Singh S, Liu Q (2013) Comparative profiling of miRNA expression in developing seeds of high linoleic and high oleic safflower (Carthamus tinctorius L.) plants. Frontiers in. Plant Sci 4(489)
Chen D, Yan W, Fu LY, Kaufmann K (2018) Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat Commun 9(1):1–3
Choudhary S, Sethy NK, Shokeen B, Bhatia S (2009) Development of chickpea EST–SSR markers and analysis of allelic variation across related species. Theor Appl Genet 118:591–608. https://doi.org/10.1007/s00122-008-0923-z
Creighton CJ, Reid JG, Gunaratne PH (2009) Expression profiling of microRNAs by deep sequencing. Brief Bioinform 10(5):490–497. https://doi.org/10.1093/bib/bbp019
Czech B, Hannon GJ (2011) Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12(1):19–31
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39(suppl_2):W155–W159. https://doi.org/10.1093/nar/gkr319
Duan H, Lu X, Lian C, An Y, Xia X, Yin W (2016) Genome-wide analysis of microRNA responses to the phytohormone abscisic acid in Populus euphratica. Front Plant Sci 7:1184
Dussert S, Serret J, Bastos-Siqueira A, Morcillo F, Déchamp E, Rofidal V, Lashermes P, Etienne H, Joët T (2018) Integrative analysis of the late maturation programme and desiccation tolerance mechanisms in intermediate coffee seeds. J Exp Bot 69(7):1583–1597
Garg R, Sahoo A, Tyagi AK, Jain M (2010) Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem Biophys Res Commun 396(2):283–288
German MA, Luo S, Schroth G, Meyers BC, Green PJ (2009) Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4(3):356–362
Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18(8):1873–1886. https://doi.org/10.1105/tpc.105.037192
Gomes CP, Cho JH, Hood LE, Franco OL, Pereira RW, Wang K (2013) A review of computational tools in microRNA discovery. Front Genet 4:81. https://doi.org/10.3389/fgene.2013.00081
Hofacker IL (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31(13):3429–3431
Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138(19):4117–4129
Jain M, Chevala VN, Garg R (2014) Genome-wide discovery and differential regulation of conserved and novel microRNAs in chickpea via deep sequencing. J Exp Bot 65(20):5945–5958
Jiao Y, Song W, Zhang M, Lai J (2011) Identification of novel maize miRNAs by measuring the precision of precursor processing. BMC Plant Biol 11(1):141. https://doi.org/10.1186/1471-2229-11-141
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42(6):541–544
Jin Y, Liu L, Hao X, Harry DE, Zheng Y, Huang T, Huang J (2019) Unravelling the microRNA-mediated gene regulation in developing Pongamia seeds by high-throughput small RNA profiling. Int J Mol Sci 20(14):3509
Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G (2016) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res Oct 23:gkw982
Khandal H, Parween S, Roy R, Meena MK, Chattopadhyay D (2017) MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci Rep 7:4632. https://doi.org/10.1038/s41598-017-04906-z
Khemka N, Singh VK, Garg R, Jain M (2016) Genome-wide analysis of long intergenic non-coding RNAs in chickpea and their potential role in flower development. Sci Rep 6:33297
Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta (BBA)-Gene Regulatory Mechanisms 1819(2):137–148
Kohli D, Joshi G, Deokar AA, Bhardwaj AR, Agarwal M, Katiyar-Agarwal S, Srinivasan R, Jain PK (2014) Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PloS one 9(10):e108851
Koroban NV, Kudryavtseva AV, Krasnov GS, Sadritdinova AF, Fedorova MS, Snezhkina AV, Bolsheva NL, Muravenko OV, Dmitriev AA, Melnikova NV (2016) The role of microRNA in abiotic stress response in plants. Mol Biol 50(3):337–343
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47(D1):D155–D162
Kukurba KR, Montgomery SB (2015) RNA sequencing and analysis. Cold Spring Harbor Protocols. 2015(11):pdb-top084970.
Lee WC, Lu SH, Lu MH, Yang CJ, Wu SH, Chen HM (2015) Asymmetric bulges and mismatches determine 20-nt microRNA formation in plants. RNA Biol 12(9):1054–1066
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Li T, Ma L, Geng Y, Hao C, Chen X, Zhang X (2015) Small RNA and degradome sequencing reveal complex roles of miRNAs and their targets in developing wheat grains. PloS one 10(10):e0139658
Liu X, Huang J, Wang Y, Khanna K, Xie Z, Owen HA, Zhao D (2010) The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J 62(3):416–428
Liu H, Qin C, Chen Z, Zuo T, Yang X, Zhou H, Xu M, Cao S, Shen Y, Lin H, He X (2014) Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics 15(1):25
Liu N, Wu S, Li Z, Khan AQ, Hu H, Zhang X, Tu L (2020) Repression of microRNA 160 results in retarded seed integument growth and smaller final seed size in cotton. Crop J 8:602–612
Liu MY, Wu XM, Long JM, Guo WW (2017) Genomic characterization of miR156 and SQUAMOSA promoter binding protein-like genes in sweet orange (Citrus sinensis). Plant Cell, Tissue Organ Cult (PCTOC) 130(1):103–116
Ma X, Zhang X, Zhao K, Li F, Li K, Ning L, He J, Xin Z, Yin D (2018) Small RNA and degradome deep sequencing reveals the roles of microRNAs in seed expansion in peanut (Arachis hypogaea L.). Front Plant Sci 9(349)
Malik N, Dwivedi N, Singh AK, Parida SK, Agarwal P, Thakur JK, Tyagi AK (2016) An integrated genomic strategy delineates candidate mediator genes regulating grain size and weight in rice. Sci Rep 6(1):1–2
Malovichko YV, Shtark OY, Vasileva EN, Nizhnikov AA, Antonets KS (2020) Transcriptomic insights into mechanisms of early seed maturation in the garden pea (Pisum sativum L.). Cells 9(3):779
Mao HD, Yu LJ, Li ZJ, Yan Y, Han R, Liu H, Ma M (2016) Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 6:1–2
Martin RC, Martínez-Andújar C, Nonogaki H (2012) Role of miRNAs in seed development. In: Sunkar R (ed) MicroRNAs in Plant Development and Stress Responses. Springer, Berlin, Heidelberg, pp 109–121. https://doi.org/10.1007/978-3-642-27384-1_6
Meyers BC, Axtell MJ (2019) MicroRNAs in plants: Key findings from the early years. Plant Cell 31(6):1206–1207. https://doi.org/10.1105/tpc.19.00310
Miao C, Wang D, He R, Liu S, Zhu JK (2020) Mutations in MIR 396e and MIR 396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol J 18(2):491–501
Montes RA, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martínez N, Meyers BC, Green PJ, de Folter S (2014) Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun 5(1):1–5
Nodine MD, Bartel DP (2010) MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev 24(23):2678–2692
Ortiz JP, Leblanc O, Rohr C, Grisolia M, Siena LA, Podio M, Colono C, Azzaro C, Pessino SC (2019) Small RNA-seq reveals novel regulatory components for apomixis in Paspalum notatum. BMC Genomics 20(1):487
Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53(5):731
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PloS one 7(2):e30619
Peng T, Sun H, Du Y, Zhang J, Li J, Liu Y, Zhao Y, Zhao Q (2013) Characterization and expression patterns of microRNAs involved in rice grain filling. PloS one 8(1):e54148
Porta H, Rocha-Sosa M (2002) Plant lipoxygenases. Physiological and molecular features. Plant Physiol 130(1):15–21
Pradhan S, Bandhiwal N, Shah N, Kant C, Gaur R, Bhatia S (2014) Global transcriptome analysis of developing chickpea (Cicer arietinum L.) seeds. Frontiers in. Plant Sci 5(698)
Pradhan S, Kant C, Verma S, Bhatia S (2017) Genome-wide analysis of the CCCH zinc finger family identifies tissue specific and stress responsive candidates in chickpea (Cicer arietinum L.). PloS one 12(7):e0180469. https://doi.org/10.1371/journal.pone.0180469
Raboy V, Dickinson DB (1984) Effect of phosphorus and zinc nutrition on soybean seed phytic acid and zinc. Plant Physiol 75(4):1094–1098
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25(7):2383–2399. https://doi.org/10.1105/tpc.113.113159
Schreiber AW, Shi B-J, Huang C-Y, Langridge P, Baumann U (2011) Discovery of barley miRNAs through deep sequencing of short reads. BMC Genomics 12:129. https://doi.org/10.1186/1471-2164-12-129
Shen W, Yao X, Ye T, Ma S, Liu X, Yin X, Wu Y (2018) Arabidopsis Aspartic Protease ASPG1 Affects Seed Dormancy, Seed Longevity and Seed Germination. Plant Cell Physiol 59:1415–1431
Sheng L, Chai W, Gong X, Zhou L, Cai R, Li X, Zhao Y, Jiang H, Cheng B (2015) Identification and characterization of novel maize miRNAs involved in different genetic background. Int J Biol Sci 11(7):781–793
Shi Y, Mandal R, Singh A, Pratap Singh A (2020) Legume lipoxygenase: Strategies for application in food industry. Legum Sci 2(3):e44
Shunmugam AS, Bock C, Arganosa GC, Georges F, Gray GR, Warkentin TD (2015) Accumulation of phosphorus-containing compounds in developing seeds of low-phytate pea (Pisum sativum L.) mutants. Plants 4(1):1–26
Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, Chen E (2016) OsSPL13 controls grain size in cultivated rice. Nature Genet 48(4):447–456
Song X, Li Y, Cao X, Qi Y (2019) MicroRNAs and their regulatory roles in plant–environment interactions. Annu Rev Plant Biol 70:489–525
Srivastava S, Zheng Y, Kudapa H, Jagadeeswaran G, Hivrale V, Varshney RK, Sunkar R (2015) High throughput sequencing of small RNA component of leaves and inflorescence revealed conserved and novel miRNAs as well as phasiRNA loci in chickpea. Plant Sci 235:46–57
Tang M, Bai X, Niu LJ, Chai X, Chen MS, Xu ZF (2018) miR172 regulates both vegetative and reproductive development in the perennial woody plant Jatropha Curcas. Plant Cell Physiol 59(12):2549–2563
Tang X, Bian S, Tang M, Lu Q, Li S, Liu X, Tian G, Nguyen V, Tsang EW, Wang A, Rothstein SJ (2012) MicroRNA–mediated repression of the seed maturation program during vegetative development in Arabidopsis. PLoS Genet 8(11):e1003091
Tang J, Chu C (2017) MicroRNAs in crop improvement: fine-tuners for complex traits. Nat Plants 3(7):1–1
Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E (2008) Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J 53(1):42–52
Tian B, Wang S, Todd TC, Johnson CD, Tang G, Trick HN (2017) Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genomics 18:572. https://doi.org/10.1186/s12864-017-3963-4
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3(1):1–2. https://doi.org/10.1186/1746-4811-3-12
Verma S, Bhatia S (2019) Analysis of genes encoding seed storage proteins (SSPs) in chickpea (Cicer arietinum L.) reveals co-expressing transcription factors and a seed-specific promoter. Functional Integr Genomics 19(3):373–390
Wang J, Mei J, Ren G (2019) Plant microRNAs: biogenesis, homeostasis, and degradation. Front Plant Sci 10:360
Wang WQ, Wang J, Wu YY, Li DW, Allan AC, Yin XR (2020) Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytologist 225(4):1618–1634
Wei W, Li G, Jiang X, Wang Y, Ma Z, Niu Z, Wang Z, Geng X (2018) Small RNA and degradome profiling involved in seed development and oil synthesis of Brassica napus. PloS one 13(10):e0204998
Wollmann H, Mica E, Todesco M, Long JA, Weigel D (2010) On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137(21):3633–3642
Yang X, Li L (2011) miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 27(18):2614–2615
Yang T, Guo L, Ji C, Wang H, Wang J, Zheng X, Xiao Q, Wu Y (2020) The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. The Plant Cell
Yang T, Wang Y, Teotia S, Zhang Z, Tang G (2018) The making of leaves: how small RNA networks modulate leaf development. Front Plant Sci 9:824
Yao JL, Tomes S, Xu J, Gleave AP (2016) How microRNA172 affects fruit growth in different species is dependent on fruit type. Plant Signal Behav 11(4):417–427
Yu JY, Zhang ZG, Huang SY, Han X, Wang XY, Pan WJ, Qin HT, Qi HD, Yin ZG, Qu KX, Zhang ZX (2019) Analysis of miRNAs Targeted Storage Regulatory Genes during Soybean Seed Development Based on Transcriptome Sequencing. Genes 10(6):408
Zhang Z, Dong J, Ji C, Wu Y, Messing J (2019) NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci 116(23):11223–11228
Zhang X, Xie S, Han J, Zhou Y, Liu C, Zhou Z, Wang F, Cheng Z, Zhang J, Hu Y, Hao Z (2019) Integrated transcriptome, small RNA, and degradome analysis reveals the complex network regulating starch biosynthesis in maize. BMC Genomics 20(1):574
Zhang B, Zhang X, Liu G, Guo L, Qi T, Zhang M, Li X, Wang H, Tang H, Qiao X, Pei W (2018) A combined small RNA and transcriptome sequencing analysis reveal regulatory roles of miRNAs during anther development of Upland cotton carrying cytoplasmic male sterile Gossypium harknessii (D2) cytoplasm. BMC Plant Biol 18(1):242
Zhao Y, Wang S, Wu W, Li L, Jiang T, Zheng B (2018) Clearance of maternal barriers by paternal miR159 to initiate endosperm nuclear division in Arabidopsis. Nat Commun 9(1):1–1
Zhou X, Khare T, Kumar V (2020) Recent trends and advances in identification and functional characterization of plant miRNAs. Acta Physiol Plantarum 42(2):25
Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145(2):242–256
Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9(1):149
Acknowledgements
SP and SV acknowledge the award of research fellowship from the Department of Biotechnology, Govt. of India. AC acknowledges the award of a research fellowship from the CSIR, Govt. of India. The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources. The authors also thank Dr. Swarup Parida, NIPGR, New Delhi, India for providing the chickpea seeds for LPC and HPC genotypes.
Funding
This work was funded by the Department of Biotechnology, Government of India, under the Challenge Programme on Chickpea Functional Genomics (grant number: BT/AGR/CG-PhaseII/01/2014).
Author information
Authors and Affiliations
Contributions
SP, SV, AC, and SB were involved in the designing and execution of the work. SP, SV, and AC conducted all the experiments, analyzed data, and prepared the manuscript draft. SB corrected the manuscript and gave the final approval for the version to be published.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pradhan, S., Verma, S., Chakraborty, A. et al. Identification and molecular characterization of miRNAs and their target genes associated with seed development through small RNA sequencing in chickpea. Funct Integr Genomics 21, 283–298 (2021). https://doi.org/10.1007/s10142-021-00777-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-021-00777-w