Abstract
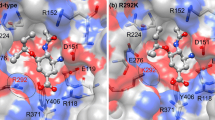
The outbreak of avian influenza virus H5N1 has raised a global concern because of its high virulence and mutation rate. Although two classes of antiviral drugs, M2 ion channel protein inhibitors and neuraminidase inhibitors, are expected to be important in controlling the early stages of a potential pandemic. Different strains of influenza viruses have differing degrees of resistance against the antivirals. In order to analyze the detailed information on the viral resistance, molecular dynamics simulations were carried out for the neuraminidase (NA) complex with oseltamivir. The carboxylate of Glu276 of H252Y NA faces toward the O-ethyl-propyl group of oesltamivir, Glu276 of wild-type NA adopts a conformation pointing away from the oesltamivir. τ2 and τ3 torsional angles fluctuation of the oesltamivir are relatively high for the H252Y mutant NA complex. In addition, there are fewer hydrogen bonds between the oesltamivir and H252Y mutation NA. The results show that H252Y mutation NA has high resistance against the drug.






Similar content being viewed by others
References
Enserink M (2005) Avian influenza: pandemic influenza: global update. Science 309:370–371
de Jong MD, Hien TT (2006) Avian influenza A (H5N1). J Clin Virol 35:2–13
Colman PM (1994) Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci 3:1687–1696
Matrosovich MN, Matrosovich TY, Gray T et al (2004) Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78:12665–12667
Douglas RG Jr (1990) Prophylaxis and treatment of influenza. N Engl J Med 322:443–450
Wintermeyer SM, Nahata MC (1995) Rimantadine: a clinical perspective. Ann Pharmacother 29:299–310
Pinto LH, Holsinger LJ, Lamb RA (1992) Influenza virus M2 protein has ion channel activity. Cell 69:517–528
Hay AJ et al (1993) In: Hannoun CE (ed) Options for the control of influenza virus II. Excerpta Medica, Amsterdam, pp 281–288
Jefferson T, Demicheli V, Rivetti D et al (2006) Antivirals for influenza in healthy adults: systematic review. Lancet 367:303–313
Gubareva LV, Kaiser L, Matrosovich MN et al (2001) Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis 183:523–531
McKimm-Breschkin JL (2000) Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res 47:1–17
Moscona A (2005) Oseltamivir resistance-disabling our influenza defenses. N Engl J Med 353:2633–2636
Ferraris O, Lina B (2008) Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J Clin Virol 41:13–19
Adcock SA, McCammon JA (2006) Molecular dynamics: survey of methods for simulating the activity of proteins. Chem Rev 106:1589–1615
van Gusteren WF, Bakowies D, Baron R et al (2006) Biomolecular modeling: goals, problems, perspectives. Angew Chem Int Ed Eng 45:4064–4092
Ravna AW, Sylte I, Dahl SG (2009) Structure and localisation of drug binding sites on neurotransmitter transporters. J Mol Model 15:1155–1164
Masukawa KM, Kollman PA, Kuntz ID (2003) Investigation of Neuraminidase Substrate Recognition Using Molecular Dynamics and Free Energy Calculations. J Med Chem 46:5628–5637
Malaisree M, Rungrotmongkol T, Decha P et al (2008) Understanding of known drug-target interactions in the catalytic pocket of neuraminidase subtype N1. Proteins 17:1908–1918
Malaisree M, Rungrotmongkol T, Nunthaboot N et al (2009) Source of oseltamivir resistance in avian influenza H5N1 virus with the H274Y mutation. Amino Acids 37:725–732
Udommaneethanakit T, Rungrotmongkol T, Bren U et al (2009) Dynamic behavior of avian influenza a virus neuraminidase subtype H5N1 in complex with oseltamivir, zanamivir, peramivir, and their phosphonate analogues. J Chem Inf Model 49:2323–2332
Amaro RE, Minh DD, Cheng LS et al (2007) Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J Am Chem Soc 129:7764–7765
Landon MR, Amaro RE, Baron R et al (2008) Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem Biol Drug Des 71:106–116
Rameix-Welti MA, Agou F, Buchy P et al (2006) Natural variation can significantly alter the sensitivity of influenza A (H5N1) viruses to oseltamivir. Antimicrob Agents Chemother 50:3809–3815
Govorkova EA, Ilyushina NA, Boltz DA, Douglas A, Yilmaz N, Webster RG (2007) Efficacy of oseltamivir therapy in ferrets inoculated with different clades of H5N1 influenza virus. Antimicrob Agents Chemother 51:1414–1424
McKimm-Breschkin JL, Selleck PW, Usman TB et al (2007) Reduced sensitivity of influenza A (H5N1) to oseltamivir. Emerging Infectious Diseases 13:1354–1357
Russell RJ, Haire LF, Stevens DJ et al (2006) The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49
Frisch MJ, Trucks GW, Schlegel HB et al (1998) Gaussian98, Revision A.7. Gaussian Inc, Pittsburgh, PA
Bayly C, Cieplak P, Cornell W et al (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J Phys Chem 97:10269–10280
Wang J, Wang W, Kollman PA et al (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25(2):247–260
Jorgensen WL, Chandrasekhar J, Madura J et al (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Duan Y, Wu C, Chowdhury S, Lee MC et al (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Lee MC, Duan Y (2004) Distinguish protein decoys using a scoring function based on AMBER force field, short molecular dynamics simulations and the generalized born solvent model. Proteins 55(3):620–634
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174
Darden T, York D, Pedersen L (1993) Particle mesh Ewald—an Nlog(N) method for Ewald sums in large systems. J Chem Phy 98:10089–10092
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 60873103), and supported by the Key Project of Natural Science Foundation of China (No. 30830090), and supported by Program for New Century Excellent Talents in University (No.NCET-06-0780), and supported by Visitorg Scholar Foundation of Key Laboratory of Biorheological Science and Technology (No. 2009BST03).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shu, M., Lin, Z., Zhang, Y. et al. Molecular dynamics simulation of oseltamivir resistance in neuraminidase of avian influenza H5N1 virus. J Mol Model 17, 587–592 (2011). https://doi.org/10.1007/s00894-010-0757-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0757-x