Abstract
Fluorophore 1,8-naphthilamide was linked to 2-bromoacridine through an ethylenediamine spacer using a succinct synthetic route to give a bromoacridine-linked, bifunctional fluorophore conjugate for the detection of triplex DNA. Acridine is well known to intercalate into duplex DNA whereas introduction of a bulky bromine atom at position C2 redirects specificity for triplex over duplex DNA. In this work, photoelectron transfer assay was used to demonstrate that the synthesised 2-bromoacridine-linked fluorophore conjugate had good selectivity for the representative triplex DNA target sequence d(T*A.T)20 compared with double-stranded d(T.A)20, single-stranded dT20 or d(G/A)19 DNA sequences.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of triplex-forming oligonucleotides allows potentially sequence-specific targeting of genes in humans and in other organisms to control or alter gene expression [1]. The structural and assembly characteristics that typify DNA triplexes allow the design of DNA nanonstructures for structural applications and targeting of non-nucleic acid components [2]. Representative triplex structures are shown, whereby the Watson–Crick A.T and G.C DNA base pairs are targeted by T (Fig. 1, left) and protonated C+ (Fig. 1, right) [3].
Of the various methods that exist for the study of DNA triplexes, variable temperature UV analysis and non-denaturing polyacrylamide gel electrophoresis (PAGE) are commonly used. There are examples where stable DNA triplexes are expected to form but fail to show temperature-dependant changes in hyperchromicity thus rendering triplex formation “invisible” to UV analysis in such instances [4]. The use of non-denaturing PAGE is highly effective for both the qualitative and quantitative analyses of DNA triplexes but the use of radioactive material containing 32P and/or other radionuclides requires expertise in handling, manufacture and the safe disposal of waste to protect against negative impact on the environment. Fluorescence-based [5] approaches provide potentially less expensive, greener methods for triplex DNA analysis. The use of a bifunctional triplex DNA sensor incorporating the 1,8-naphthilamide fluorophore and a naphthoquinoline motif was examined [6] and shown to be selective for triplex over duplex and single-stranded DNA, as judged by photoelectron transfer [7] (PET) assay.
We wished to examine whether the PET assay approach could be extended to the use of the acridine conjugate 7 with the simplicity of using bromine substitution within the acridine ligand to promote recognition and binding to triplex DNA. Normally, acridine acts as a potent duplex DNA intercalator [8, 9] but the presence of bromine atom at position C2 has been shown to redirect its specificity for triplex DNA [10].
Results and discussion
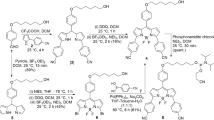
To synthesise conjugate 7, we first prepared 2-bromo-9-chloroacridine (3) (Scheme 1). N-(4-Bromophenyl)anthranilic acid (2) was prepared from 2-chlorobenzoic acid (1) by a literature modification to the Ullman synthesis [11]. The anthranilic acid 2 was then cyclised with concomitant chlorination to give the acridine 3 [12, 13].

The preparation of the 4-bromonaphthalimide 6 was achieved by reacting compound 4 with methylamine [14] to give compound 5, that was subsequently treated with ethylenediamine [15] (Scheme 2). The coupling of acridine 3 to naphthalimide 6 was successful and gave the target conjugate 7, albeit it in very low yield. The structures of conjugate 7 and the intermediates were consistent with their spectroscopic properties. Representative spectroscopic data are given in the Supplementary Material.

DNA sequences dT20, dA20 and the mixed G/A sequence were prepared by solid phase synthesis using phosphoramidites on a Beckman Oligo 1000 DNA synthesiser according to the manufacturer’s protocol. The oligonucleotide products were purified by reversed phase HPLC. Representative HPLC chromatograms are given in the Supplementary Material.
The increase in fluorescence enhancement shown by conjugate 7 in the presence of triplex DNA, correlated with the increase in concentration of the DNA triplex, as evidenced by the PET assay results summarised in Fig. 2. To examine the selectivity for triplex DNA, conjugate 7 (1 μM) was separately added to the same total concentration (40 μM) of triplex (TAT), duplex (AT), two types of single-stranded DNA (T and G/A), and the fluorescence enhancements were examined by PET assay (Fig. 3). Conjugate 7 was not specific for triplex DNA but it did, however, show marked selectivity for triplex DNA compared with the duplex DNA and single-stranded DNA sequences.
Emission spectra (excitation at 457 nm) of conjugate 7 (1 μM) in the presence of various concentrations of DNA triplex d(T*A.T)20 ranging from 0, 5, 10, 20, 40, 60, 80, and 100 μM measured in 25 mM Tris.HCl buffer containing 10 mM MgCl2, 100 mM NaCl, and 0.5 mM spermine at pH 7. Fluorescence is given in relative fluorescence units (RFU)
Fluorescence response of conjugate 7 (1 μM) to various nucleic acid samples (40 μM) in 25 mM Tris.HCl buffer containing 10 mM MgCl2, 100 mM NaCl, and 0.5 mM spermine at pH 7. TAT represents the triplex formed from 2 molar equivalents of dT20 and 1 molar equivalent of dA20; AT represents the duplex formed from 1 molar equivalent of dT20 and 1 molar equivalent of dA20; T represents single-stranded dT20; G/A represents the 19-mer sequence 5ʹ-dAGAGAGGAGAGAAGAGGAG; none represents the fluorescence response in the absence of nucleic acid
Conclusion
We have shown conjugate 7 to be selective for triplex DNA compared with duplex or single-stranded DNA. Although its discriminatory properties were not superior to the naphthoquinoline conjugate described by Lu and co-workers [6], its concise synthesis should allow access to further substitution patterns in the acridine portion to potentially fine-tune the selectivity for triplex DNA.
Experimental
NMR spectra were recorded on a Bruker AC-250 spectrometer in CDCl3 or DMSO-d6. The chemical shift values are expressed as δ values (ppm) down field with residual protons of the solvents (CDCl3, δ = 7.26 ppm; DMSO-d6, δ = 2.49 ppm) as internal standards. Mass spectrometric analyses were performed in FAB + mode using a VG AutoSpec instrument. Infrared spectra were recorded using a Mattson Galaxy 2020 FT-IR Spectrophotometer. Melting points were measured on a Gallenkamp Electrothermal Digital apparatus. Fluorescence spectra were recorded on a Spectramax Gemini XS dual-scanning microplate spectrofluorometer. Oligonucleotides were prepared on a Beckman Oligo 1000 DNA synthesiser following the manufacturer’s protocol using commercially available reagents (LINK Technologies). Purification of oligonucleotides was performed by semi-preparative reversed phase HPLC. The procedures for preparation of known compounds 2 [11], 3 [12, 13], 5 [14], 6 [15] and their analytical and spectroscopic data are given in the Supplementary Material.
6-[2-[(2-Bromoacridin-9-yl)amino]ethylamino]-2-methylbenzo[de]isoquinoline-1,3-dione (7, C28H21BrN4O2)
To an oven dried two-necked round bottom flask (100 cm3) was added 153 mg 2-bromo-9-chloroacridine (3, 0.52 mmol) and 983 mg phenol (10.46 mmol) under an argon atmosphere and the flask heated (80 °C) until the acridine had dissolved. The 1,8-naphthalimide 6 (128 mg, 0.48 mmol) was dissolved in the minimum of dry DMF, added to the reaction flask and the reaction mixture heated (110 °C) with stirring (2 h). The product mixture was then allowed to cool (rt) before addition of chilled ethanol (3 cm3). After refrigeration (overnight), the precipitate was collected by Buchner filtration and dried under vacuum to give the product (37 mg, 13%) as an orange solid. TLC: Rf = 0.78 (acetone-MeOH 20:1); IR: \(\overline{V}\) = 3379, 3246, 3067, 2934, 1678, 1638, 1572, 1475, 1359, 1277, 1124, 756 cm−1; 1H NMR: δ = 9.37 (1H, s, NH), 8.75 (1H, br s, NH), 8.43–8.31 (2H, m, 2 × CH), 8.05 (1H, d, J = 8.8 Hz, CH), 7.89–7.75 (2H, m, 2 × CH), 7.49–7.35 (2H, m, 2 × CH), 7.15 (2H, t, J = 7.5 Hz, 2 × CH), 6.81–6.72 (3H, m, 3 × CH), 4.50 (2H, br s, CH2), 3.94 (2H, br s, CH2), 3.39 (3H, s, CH3) ppm; 13C NMR: δ = 158.0, 150.1, 141.5, 137.6, 128.5, 125.9, 124.2, 123.2, 121.2, 121.1, 120.3, 118.4, 108.2, 104.2, 47.3, 42.5, 26.3 ppm; MS (APCI−): m/z = 523, 525; calcd for C28H21BrN4O2 ([M-H]−) 523.0770, found 523.0792.
References
Mayer A, Leumann CJ (2007) Eur J Org Chem 2007:4038
Chandrasekaran AR, Rusling DA (2018) Nucleic Acids Res 46:1021
Rusling DA, Fox KR (2014) Nucleic Acids Res 67:123
Parel SP, Leumann CJ (2001) Nucleic Acids Res 29:2260
Kanamori T, Masaki Y, Oda Y, Ohzeki H, Ohkubo A, Sekine M, Seio K (2019) Org Biomol Chem 17:2077
Lu E, Peng X, Song F, Fan J (2005) Bioorg Med Chem Lett 1:255
Liu Q, Chang Liu C, Chang L, He S, Lu Y, Zeng X (2014) RSC Adv 4:14361
Bhaduri S, Ranjan N, Arya DP (2018) Beilstein J Org Chem 14:1051
Keppler MD, McKeen CM, Zegrocka O, Strekowski L, Brown T, Fox KR (1999) Biochim Biophys Acta 1447:137
Strekowski L, Hojjat M, Wolinska E, Parker AN, Paliakov E, Gorecki T, Tanious FA, Wilson WD (2005) Bioorg Med Chem Lett 15:1097
Sharples D (1982) J Pharm Pharmacol 34:681
Robin M, Faure R, Périchaud A, Galy JP (2002) Synth Commun 32:981
Roubaud G, Faure R, Galy JP (2003) Magn Reson Chem 41:549
Magalhães JL, Pereira RV, Triboni ER, Filho PB, Gehlen MH, Nart FC (2006) J Photochem Photobiol A 183:165
Fan J, Peng X, Wu Y, Lu E, Hou J, Zhang H, Zhang R, Fu X (2005) J Lumin 114:125
Acknowledgements
We thank Karen Farrow (Aston University) for technical expertise in mass spectrometric analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharma, S.K., Fraser, W. Selectivity of a bromoacridine-containing fluorophore for triplex DNA. Monatsh Chem 152, 1013–1016 (2021). https://doi.org/10.1007/s00706-021-02816-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00706-021-02816-5