Abstract
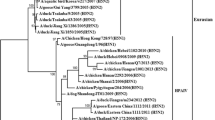
Aquatic birds are known to be a reservoir for the most common influenza A viruses (IAVs). In the annual surveillance program, we collected the feces of migratory birds for the detection of IAVs in South Korea in November 2016. A novel reassorted H3N3 avian influenza virus (AIV) containing genes from viruses of wild and domestic birds was identified and named A/aquatic bird/South Korea/sw006/2016(H3N3). The polymerase basic 2 (PB2) and non-structural (NS) genes of this isolate are most closely related to those of wild-bird-origin AIV, while the polymerase basic 1 (PB1), polymerase acidic (PA), hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), and matrix (M) genes are most closely related to those of domestic-bird-origin AIV. A/aquatic bird/South Korea/sw006/2016 contains PA, NP, M, and NS genes were most closely related to those of AIV subtype H4 and PB2, PB1, and HA genes that are most closely related to those of AIV subtype H3N8, while the NA gene was most closely related to those of subtype H10, which was recently detected in humans in China. These results suggest that novel reassortment of AIV strains occurred due to interaction between wild and domestic birds. Hence, we emphasize the need for continued surveillance of avian influenza virus in bird populations.

Similar content being viewed by others
References
Yoon SW, Webby RJ, Webster RG (2014) Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol 385:359–375
Aligne CA (2016) Overcrowding and mortality during the influenza pandemic of 1918. Am J Public Health 106:642–644
Peiris JS, Poon LL, Guan Y (2009) Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J Clin Virol 45:169–173
Viboud C, Simonsen L, Fuentes R et al (2016) Global mortality impact of the 1957–1959 influenza pandemic. J Infect Dis 213:738–745
Wendel I, Rubbenstroth D, Doedt J et al (2015) The avian-origin PB1 gene segment facilitated replication and transmissibility of the H3N2/1968 pandemic influenza virus. J Virol 89:4170–4179
Chen H, Yuan H, Gao R et al (2014) Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet (London, England) 383:714–721
Scholtissek C, Rohde W, Von Hoyningen V et al (1978) On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13–20
Zhou NN, Senne DA, Landgraf JS et al (2000) Emergence of H3N2 reassortant influenza A viruses in North American pigs. Vet Microbiol 74:47–58
Crawford PC, Dubovi EJ, Castleman WL et al (2005) Transmission of equine influenza virus to dogs. Science 310:482–485
Song D, Kang B, Lee C et al (2008) Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 14:741–746
Su S, Cao N, Chen J et al (2012) Complete genome sequence of an avian-origin H3N2 canine influenza A virus isolated in farmed dogs in southern China. J Virol 86:10238
Spackman E, Senne DA, Myers TJ et al (2002) Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40:3256–3260
Yoo HS, Eah JY, Kim JS et al (2006) DNA barcoding Korean birds. Mol Cells 22:323–327
Hoffmann E, Stech J, Guan Y et al (2001) Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289
Matrosovich MN, Gambaryan AS, Teneberg S et al (1997) Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234
Gabriel G, Dauber B, Wolff T et al (2005) The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci USA 102:18590–18595
Baek YH, Song MS, Lee EY et al (2015) Profiling and characterization of influenza virus N1 strains potentially resistant to multiple neuraminidase inhibitors. J Virol 89:287–299
Hay AJ, Wolstenholme AJ, Skehel JJ et al (1985) The molecular basis of the specific anti-influenza action of amantadine. EMBO J 4:3021–3024
Wang J, Wu Y, Ma C et al (2013) Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc Natl Acad Sci USA 110:1315–1320
Fan S, Deng G, Song J et al (2009) Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 384:28–32
Hale BG, Randall RE, Ortin J et al (2008) The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376
Obenauer JC, Denson J, Mehta PK et al (2006) Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580
Choi JG, Kang HM, Kim MC et al (2012) Genetic relationship of H3 subtype avian influenza viruses isolated from domestic ducks and wild birds in Korea and their pathogenic potential in chickens and ducks. Vet Microbiol 155:147–157
Wu H, Wu N, Peng X et al (2014) Molecular characterization and phylogenetic analysis of H3 subtype avian influenza viruses isolated from domestic ducks in Zhejiang Province in China. Virus Genes 49:80–88
Callan RJ, Early G, Kida H et al (1995) The appearance of H3 influenza viruses in seals. J Gen Virol 76(Pt 1):199–203
Karasin AI, West K, Carman S et al (2004) Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol 42:4349–4354
Song MS, Oh TK, Moon HJ et al (2008) Ecology of H3 avian influenza viruses in Korea and assessment of their pathogenic potentials. J Gen Virol 89(Pt 4):949–957
Olsen B, Munster VJ, Wallensten A et al (2006) Global patterns of influenza a virus in wild birds. Science 312:384–388
Funding
This work was supported by grants from the KRIBB Initiative program, the BioNano Health-Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as Global Frontier Project (Grant No. H-GUARD 2013M3A6B2078954) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HD16A1487).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study did not include any experiments with human or animal subjects.
Additional information
Handling Editor: Ayato Takada.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Le, T.B., Kim, H.K., Le, H.Y. et al. Complete genome sequence of a novel reassortant H3N3 avian influenza virus. Arch Virol 164, 2881–2885 (2019). https://doi.org/10.1007/s00705-019-04386-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04386-8