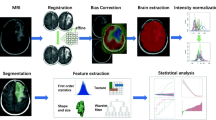
Abstract
Objective
To develop a nomogram based on MRI radiomics and clinical features for preoperatively predicting H3K27M mutation in pediatric high-grade gliomas (pHGGs) with a midline location of the brain.
Methods
The institutional database was reviewed to identify patients with pHGGs with a midline location of the brain who underwent tumor biopsy with preoperative MRI scans between June 2016 and June 2021. A total of 107 patients with pHGGs, including 79 patients with H3K27M mutation, were consecutively included and randomly divided into training and test sets. Radiomics features were extracted from fluid-attenuated inversion recovery (FLAIR), diffusion-weighted (DW) and post-contrast T1-weighted images, and apparent diffusion coefficient (ADC) maps. The minimum redundancy maximum relevance (MRMR) and least absolute shrinkage and selection operator (LASSO) logistic regression were performed for radiomics signature construction. Clinical and radiological features were analyzed to select clinical predictors. A nomogram was then developed by incorporating the radiomics signature and selected clinical predictors.
Results
Nine radiomics features were selected to construct the radiomics signature, which showed a favorable discriminatory ability in training and test sets with an area under the curve (AUC) of 0.95 and 0.92, respectively. Ring enhancement was identified as an independent clinical predictor (p < 0.01). The nomogram, constructed with radiomics signature and ring enhancement, showed good calibration and discrimination in training and testing sets (AUC: 0.95 and 0.90 respectively).
Conclusions
The nomogram which combined radiomics signature and ring enhancement had a satisfactory ability to predict H3K27M mutation in pHGGs with a midline of the brain.
Key Points
• Conventional MRI features were not powerful enough to predict H3K27M mutation status in pediatric high-grade gliomas (pHGGs) with a midline location of the brain.
• An MRI-based radiomics signature showed satisfactory ability to predict H3K27M mutation status of pHGGs located in the midline of the brain.
• Associating the radiomics signature with clinical factors improved predictive performance.





Similar content being viewed by others
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AIC:
-
Akaike’s Information Criterion
- ANT:
-
Advanced Normalization Tools
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- cT1W:
-
Post-contrast T1-weighted
- DMG:
-
Diffuse midline glioma
- DW:
-
Diffusion-weighted
- DWI:
-
Diffusion-weighted images
- FLAIR:
-
Fluid-attenuated inversion recovery
- KPS:
-
Karnofsky performance status
- LASSO:
-
Least absolute shrinkage and selection operator
- MRI:
-
Magnetic resonance imaging
- MRMR:
-
Maximum relevance minimum redundancy
- pHGGs:
-
Pediatric high-grade gliomas
- Radscore:
-
Radiomic score
- ROC:
-
Receiver operator characteristic
- ROI:
-
Region of interest
- SMOTE:
-
Synthetic minority oversampling technique
- T1W:
-
T1-weighted
- WHO:
-
World Health Organization
References
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Louis DN, Giannini C, Capper D et al (2018) cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol 135:639–642
Khuong-Quang D-A, Buczkowicz P, Rakopoulos P et al (2012) K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124:439–447
Bozkurt SU, Dagcinar A, Tanrikulu B et al (2018) Significance of H3K27M mutation with specific histomorphological features and associated molecular alterations in pediatric high-grade glial tumors. Childs Nerv Syst 34:107–116
Karremann M, Gielen GH, Hoffmann M et al (2018) Diffuse high-grade gliomas with H3 K27M mutations carry a dismal prognosis independent of tumor location. Neuro Oncol 20:123–131
Ryall S, Krishnatry R, Arnoldo A et al (2016) Targeted detection of genetic alterations reveal the prognostic impact of H3K27M and MAPK pathway aberrations in paediatric thalamic glioma. Acta Neuropathol Commun 4:93
Ochs K, Ott M, Bunse T et al (2017) K27M-mutant histone-3 as a novel target for glioma immunotherapy. Oncol Immunol 6(7):e1328340
Filbin MG, Tirosh I, Hovestadt V et al (2018) Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360:331–335
Thust SC, Heiland S, Falini A et al (2018) Glioma imaging in Europe: a survey of 220 centres and recommendations for best clinical practice. Eur Radiol 28:3306–3317
Aboian MS, Solomon DA, Felton E et al (2017) Imaging characteristics of pediatric diffuse midline gliomas with histone H3 K27M mutation. AJNR Am J Neuroradiol 38:795–800
Qiu T, Chanchotisatien A, Qin Z et al (2020) Imaging characteristics of adult H3 K27M-mutant gliomas. J Neurosurg 133:1662–1670
Forghani R, Savadjiev P, Chatterjee A et al (2019) Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J 17:995–1008
Galm BP, Martinez-Salazar EL, Swearingen B et al (2018) MRI texture analysis as a predictor of tumor recurrence or progression in patients with clinically non-functioning pituitary adenomas. Eur J Endocrinol 179:191–198
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Dang M, Lysack JT, Wu T et al (2015) MRI Texture analysis predicts p53 status in head and neck squamous cell carcinoma. AJNR Am J Neuroradiol 36:166–170
Su X, Chen N, Sun H et al (2019) Automated machine learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro Oncol 22:393–401
Karsy M, Guan J, Cohen AL et al (2017) New molecular considerations for glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep 17:19
Braunstein S, Raleigh D, Bindra R et al (2017) Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J Neurooncol 134:541–549
Wesseling P, Capper D (2018) WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol 44:139–150
Shinohara RT, Sweeney EM, Goldsmith J et al (2014) Statistical normalization techniques for magnetic resonance imaging. NeuroImage Clin 6:9–19
Avants BB, Tustison NJ, Song G et al (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54:2033–2044
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Kramer AA, Zimmerman JE (2007) Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited*. Crit Care Med 35:2052–6
Vickers AJ, Cronin AM, Elkin EB et al (2008) Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 8:53
Boxerman JL, Zhang Z, Safriel Y et al (2018) Prognostic value of contrast enhancement and FLAIR for survival in newly diagnosed glioblastoma treated with and without bevacizumab: results from ACRIN 6686. Neuro Oncol 20:1400–1410
Radbruch A, Lutz K, Wiestler B et al (2012) Relevance of T2 signal changes in the assessment of progression of glioblastoma according to the Response Assessment in Neurooncology criteria. Neuro Oncol 14:222–229
Chen H, Hu W, He H et al (2019) Noninvasive assessment of H3 K27M mutational status in diffuse midline gliomas by using apparent diffusion coefficient measurements. Eur J Radiol 114:152–159
Aboian MS, Tong E, Solomon DA et al (2019) Diffusion characteristics of pediatric diffuse midline gliomas with histone H3–K27M mutation using apparent diffusion coefficient histogram analysis. AJNR Am J Neuroradiol 40:1804–1810
Lober RM, Cho Y-J, Tang Y et al (2014) Diffusion-weighted MRI derived apparent diffusion coefficient identifies prognostically distinct subgroups of pediatric diffuse intrinsic pontine glioma. J Neurooncol 117:175–182
Poussaint TY, Vajapeyam S, Ricci KI et al (2016) Apparent diffusion coefficient histogram metrics correlate with survival in diffuse intrinsic pontine glioma: a report from the Pediatric Brain Tumor Consortium. Neuro Oncol 18:725–734
LaViolette PS, Mickevicius NJ, Cochran EJ et al (2014) Precise ex vivo histological validation of heightened cellularity and diffusion-restricted necrosis in regions of dark apparent diffusion coefficient in 7 cases of high-grade glioma. Neuro Oncol 16:1599–1606
Tensaouti F, Ducassou A, Chaltiel L et al (2016) Prognostic and predictive values of diffusion and perfusion MRI in paediatric intracranial ependymomas in a large national study. Br J Radiol 89:20160537
Lu Y, Liu L, Luan S et al (2019) The diagnostic value of texture analysis in predicting WHO grades of meningiomas based on ADC maps: an attempt using decision tree and decision forest. Eur Radiol 29:1318–1328
Löbel U, Sedlacik J, Reddick WE et al (2011) Quantitative diffusion-weighted and dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging analysis of T2 hypointense lesion components in pediatric diffuse intrinsic pontine glioma. AJNR Am J Neuroradiol 32:315–322
Chiang J, Diaz AK, Makepeace L et al (2020) Clinical, imaging, and molecular analysis of pediatric pontine tumors lacking characteristic imaging features of DIPG. Acta Neuropathol Commun 8:57
Jansen MH, Veldhuijzen van Zanten SE, Sanchez Aliaga E et al (2015) Survival prediction model of children with diffuse intrinsic pontine glioma based on clinical and radiological criteria. Neuro Oncol 17:160–166
Hoffman LM, Veldhuijzen van Zanten SEM, Colditz N et al (2018) Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): a collaborative report from the International and European Society for Pediatric Oncology DIPG Registries. J Clin Oncol 36:1963–1972
Smirniotopoulos JG, Murphy FM, Rushing EJ et al (2007) Patterns of contrast enhancement in the brain and meninges. Radiographics 27:525–551
Liu Y, Xu X, Yin L et al (2018) Relationship between glioblastoma heterogeneity and survival time: an mr imaging texture analysis. AJNR Am J Neuroradiol 38:1695–1701
Ditmer A, Zhang B, Shujaat T et al (2018) Diagnostic accuracy of MRI texture analysis for grading gliomas. J Neurooncol 140:583–589
Gevaert O, Mitchell LA, Achrol AS et al (2014) Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273:168–174
Cohen KJ, Jabado N, Grill J (2017) Diffuse intrinsic pontine gliomas—current management and new biologic insights. Is there a glimmer of hope? Neuro Oncol 19:1025–1034
Funding
This study was supported by grants from the National Key Research and Development Program of China (No. 2017YFC0109003) and the Projects of Science and Technology Innovation of Shanghai (No. 18411952300 and No. 18411967500).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dengbin Wang, MD, PhD, the chief of Department of Radiology, Xinhua Hospital affiliated to Shanghai Jiao Tong University School of Medicine.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Shaofeng Duan kindly provided statistical advice for this manuscript.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chenqing Wu and Hui Zheng contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, C., Zheng, H., Li, J. et al. MRI-based radiomics signature and clinical factor for predicting H3K27M mutation in pediatric high-grade gliomas located in the midline of the brain. Eur Radiol 32, 1813–1822 (2022). https://doi.org/10.1007/s00330-021-08234-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-08234-9