Abstract
Myeloproliferative neoplasms (MPN), comprising essential thrombocythemia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF), are hematological disorders of the myeloid lineage characterized by hyperproliferation of mature blood cells. The prediction of the clinical course and progression remains difficult and new therapeutic modalities are required. We conducted a CD34+ gene expression study to identify signatures and potential biomarkers in the different MPN subtypes with the aim to improve treatment and prevent the transformation from the rather benign chronic state to a more malignant aggressive state. We report here on a systematic gene expression analysis (GEA) of CD34+ peripheral blood or bone marrow cells derived from 30 patients with MPN including all subtypes (ET (n = 6), PV (n = 11), PMF (n = 9), secondary MF (SMF; post-ET-/post-PV-MF; n = 4)) and six healthy donors. GEA revealed a variety of differentially regulated genes in the different MPN subtypes vs. controls, with a higher number in PMF/SMF (200/272 genes) than in ET/PV (132/121). PROGENγ analysis revealed significant induction of TNFα/NF-κB signaling (particularly in SMF) and reduction of estrogen signaling (PMF and SMF). Consistently, inflammatory GO terms were enriched in PMF/SMF, whereas RNA splicing–associated biological processes were downregulated in PMF. Differentially regulated genes that might be utilized as diagnostic/prognostic markers were identified, such as AREG, CYBB, DNTT, TIMD4, VCAM1, and S100 family members (S100A4/8/9/10/12). Additionally, 98 genes (including CLEC1B, CMTM5, CXCL8, DACH1, and RADX) were deregulated solely in SMF and may be used to predict progression from early to late stage MPN.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Classical Philadelphia chromosome-negative (Ph−) myeloproliferative neoplasms (MPN) can be subdivided into essential thrombocythemia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF). Although clinically distinct, they typically share mutations in JAK2, CALR, or MPL, or are termed triple-negative in the absence of these mutations [1,2,3]. JAK2V617F is the most abundant mutation, present in over 90% of PV, and 55–65% of ET and PMF patients [4].
Although the modified WHO diagnostic criteria include the presence of one of the abovementioned mutations as major criteria, cooperating mutations in other genes implicated in epigenetic regulation, transcriptional regulation, mRNA splicing, and signal transduction are often observed in PMF or SMF [5]. The impact of these mutations that are also associated with disease progression has been incorporated into prognostic scores such as MIPSS70 [6]. Furthermore, it was reported that some MPN courses are more dependent on non-classical driver mutations, and this might play a major role in transformation [7]. Despite specific subtypes and driver mutations, the clinical course varies among MPN patients, but the underlying mechanisms for this variability are still not fully understood.
The identification of gene expression profiles and biomarkers that enable the prediction of transition from a rather “benign” (i.e., mostly characterized on the cellular level by increased proliferation) chronic to a more malignant aggressive state (defined by the acquisition of a differentiation block or induction of fibrotic transformation) is important in order to develop targeted therapeutic strategies and prevent hematological progression to secondary myelofibrosis (SMF) or transformation to acute myeloid leukemia (AML). Among the Ph− MPN subtypes, ET and PV are considered early stage MPN. They are typically characterized by a relatively indolent disease course, as are some prefibrotic PMF. However, they all bare the risk of thromboembolic complications and eventually, progression into SMF and/or AML. Consequently, overt PMF and SMF are considered late stage MPN [8].
Treatment with the tyrosine-kinase inhibitor ruxolitinib is an example for rational therapy by targeting the essential pro-tumorigenic mutated JAK2 [9]. Ruxolitinib confers clinical benefit by reducing splenomegaly and other disease-related symptoms, but largely fails to induce complete molecular remission [10]. Therapeutic strategies in MPN are based on clinical conditions and laboratory results; only recently have molecular findings been integrated into risk progression models [11]. The identification of new targetable molecular components might result in further improvement of risk stratification as well as targeted therapeutic strategies. Genome-wide gene expression analysis (GEA) is a promising approach to resolve these targets.
Several GEA studies of patient-derived CD34+ cells have been conducted to investigate deregulated gene expression in MPN [12,13,14,15,16,17]. However, not all disease entities were included, thereby impeding a direct comparison of the different entities among themselves and/or their normal counterparts. Therefore, we conducted gene expression profiling of CD34+ purified bone marrow (BM)– or peripheral blood (PB)–derived cells from ET, PV, PMF, and SMF patients and compared them to CD34+ cells from healthy controls (HC).
Methods
Patient samples
PB and BM samples from MPN patients were obtained at the Department of Hematology, Oncology, Hemostaseology, and Stem Cell Transplantation at Uniklinik RWTH Aachen after written informed consent and approval of the local ethics committee (EK127/12). Control CD34+ cells from PB of individual healthy donors were obtained from Lonza. Unfractionated BM samples from JAK2V617F-positive untreated MPN patients within the CELL project — MPN database (MIND) — after written informed consent were provided from the Department of Internal Medicine, Hematology and Oncology of Brno University Hospital, Czech Republic. Femoral heads were obtained from the Department of Orthopedics from the Uniklinik RWTH Aachen, each after written informed consent and in compliance with the local ethics committee (EK300/13).
Next-generation sequencing
Relevant coding regions of 32 genes associated with hematologic malignancies were analyzed using an amplicon-based next-generation sequencing (NGS) panel (Truseq Custom Amplicon Kit, Illumina) as previously described [18]. Allele burdens (AB) were determined accordingly. Variants were called with a bidirectional frequency of > 5% (JAK2V617F and KITD816V > 1%) and reviewed manually.
Isolation of CD34+ cells
PB and BM MNCs (PBMCs/BMMCs) from MPN patients obtained from the Department of Hematology, Oncology, Hemostaseology, and Stem Cell Transplantation at Uniklinik RWTH Aachen were isolated with Ficoll-Paque Premium (GE Healthcare). Enrichment of the CD34+ cell population was performed by magnetic cell separation (MACS) with the CD34 MicroBead Kit (Miltenyi) following the manufacturer’s instructions. The purity of CD34+ cells was assessed by flow cytometry using CD34-FITC and CD45-APC antibodies (BD Biosciences). Samples with a purity of > 50% were used. Additional patient data is given in Supplemental Table 1–2.
RNA isolation and RT-qPCR
RNA was isolated with the RNeasy Micro kit (Qiagen). RT-qPCR was performed as described by Czech et al. [19]. Primers are listed in Supplemental Table 3.
Gene expression analysis
Genome-wide transcriptome analysis was performed using the Affymetrix HTA 2.0 platform (Thermo Fisher Scientific). 6.6 ng total RNA from each sample was prepared and hybridized to the HTA 2.0 arrays according to the WT Pico kit manual (Thermo Fisher Scientific). Batch correction was applied using sva and Combat [20]. Downstream analysis was performed in R.
Statistical analysis
Data were normalized with signal space transformation (SST) in conjunction with robust multiple-array (RMA) average normalization method (SST-RMA) with Affymetrix Expression console. Statistical analysis of RT-qPCR data was performed with the Mann–Whitney U test (GraphPad Prism 8).
Data sharing statement
Microarray data have been deposited at GEO under accession number GSE174060.
Results
Gene expression signatures in myelofibrosis patient-derived CD34+ cells show more pronounced differences compared to HCs than early stage MPN (ET/PV)
To investigate differences in the transcriptional profile of hematopoietic stem and progenitor cells (HSPCs) derived from ET, PV, PMF, and SMF patients as well as healthy donors, BM- or PB-derived CD34+ cells were subjected to GEA (Fig. 1A). The analysis included ET (n = 6), PV (n = 11), PMF (n = 9), and post-ET-MF/post-PV-MF (n = 2 each) patients, as well as HCs (n = 6; Supplemental Table 1), with JAK2V617F- (n = 23), CALR- (n = 4), MPL-mutated (n = 1), and triple-negative (n = 2) patients. The screening for MPN-associated driver and additional mutations was performed by NGS (Supplemental Table 2).
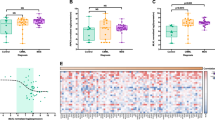
Distinct gene expression profiles of CD34+-enriched MNCs from ET, PV, PMF, and SMF patients and HC. A Experimental design of the study. Patient-derived BMMCs and PBMCs were isolated from BM aspirations or PB samples by density gradient centrifugation and enriched for the CD34+ population by magnetic cell sorting. The isolated RNA was analyzed using the Affymetrix HTA 2.0 platform and analyzed regarding gene expression, Pathway RespOnsive GENes (PROGENy), and enriched KEGG pathways and gene ontologies. B Unsupervised principal component analysis (PCA) of gene expression data between the different MPN samples and HCs. C Venn diagram of the differentially expressed genes (p < 0.05 and |FC|> 1.5). A list of all comparisons is given in Supplemental Table 4. BMMCs, bone marrow mononuclear cells; ET, essential thrombocythemia; HC, healthy controls; MNCs, mononuclear cells; PMF, primary myelofibrosis; PBMCs, peripheral blood mononuclear cells; PV, polycythemia vera; SMF, secondary myelofibrosis
Principal component analysis (PCA) revealed clustering of MPN samples and separation from HC samples (Fig. 1B). While the ET samples clustered in proximity to HCs, PV, PMF, and SMF samples were located more distantly with a broader distribution within the groups. Among the four MPN entities, we observed a continuum in dimension 2 from ET over PV, PMF, to finally SMF (Supplemental Fig. 1A). The distribution of PV, PMF, and SMF samples indicates heterogeneity between the diseases. We observed no clear clustering of the post-ET-MF/post-PV-MF samples or the different driver mutation with PCA (Supplemental Fig. 1B). Interestingly, high AB of the respective driver mutation were correlated with low PC1/PC2 values typically associated with PMF/SMF samples (Supplemental Fig. 1C) and potentially indicates beginning transformation. We analyzed differentially expressed genes compared to HCs and observed that PMF and SMF showed the highest number of differentially regulated genes (200 and 272, resp.), while PV (121) and ET (132) showed lower numbers (Fig. 1C).
Ninety-eight genes were uniquely regulated in SMF vs. HC. These genes might include potential biomarkers for progression from ET or PV to SMF, of which some will be addressed hereinafter. Furthermore, we identified 121 genes that were differentially regulated in both PMF and SMF showing some similarity. In total, 47 genes were differentially regulated in all four entities. A list of these genes and all other comparisons is provided in Supplemental Table 4. Direct comparisons between the MPN subtypes (without HCs) revealed a high degree of similarity between ET/PV and PMF/SMF with few differentially regulated genes (Supplemental Fig. 2).
To test whether the most significantly regulated genes allowed hierarchical clustering between the disease groups, a heatmap was generated by unsupervised hierarchical clustering (Fig. 2A). All HC samples clustered together. The ET samples grouped together with three PV samples that were located in between and appeared to show more PV-like expression patterns. Most PV samples grouped next to MF samples that also clustered together with three of the four SMF samples (comprising two post-ET-MF/post-PV-MF each) being adjacent to each other. Clustering analysis underlines the paradigm of “early” and “late” stage MPN. The predominant hypothesis that explains how JAK2V617F leads to three different diseases relies on the determining influence of the molecular background and the occurrence of additional mutations [21]. Increased number of concurrent mutations was shown to correlate with a PMF-like phenotype. This is reflected by our NGS analysis, as most patients carrying more than two bystander mutations were PMF patients.
Hierarchical clustering of differentially expressed genes and PROGENγ activity scores in CD34+-enriched MNCs from ET, PV, PMF, and SMF patients and HC. A Heatmap of unsupervised clustering of all 5815 differentially regulated genes of each MPN subtype vs. control samples. Allele burden of MPN-associated driver and bystander mutations is indicated by shades of gray. Samples lacking NGS data are marked with x. B Pathway RespOnsive GENes for activity inference (PROGENγ) analysis of JAK/STAT, estrogen, NF-κB, and TNFα signaling pathways in patients- or healthy donor-derived CD34+ cells, arranged by group (HC, ET, PV, PMF, and SMF), normalized in respect to HC. Pathways are restricted to significantly deregulated pathways in all MPN samples vs. HC. An overview of all pathways is given in Supplemental Fig. 3. C Two-dimensional unsupervised clustering of PROGENγ activity scores of individual HC, ET, PV, PMF, and SMF samples, normalized in respect to HC. P-values and t-statistics were calculated by t-test (* p < .05, ** p < .01, *** p < .001). ET, essential thrombocythemia; HC, healthy controls; MNCs, mononuclear cells; NGS, next-generation sequencing; PMF, primary myelofibrosis; PV, polycythemia vera; SMF, secondary myelofibrosis; #, post-ET-MF; *, post-PV-MF
NF-κB and TNFα pathways are upregulated in CD34+ cells in late stages of MPN
After elucidating single gene changes, we focused on PROGENy (Pathway RespOnsive GENes) analysis exploring the activation of cancer pathways using the top 1000 genes according to significance for the model generation [22]. Besides expected upregulation of JAK-STAT signaling, we observed strong upregulation of NF-κB and TNFα signaling compared with HCs, as well as downregulation of estrogen signaling (Fig. 2B–C). Although upregulation of these inflammatory pathways has been described [23], we could show a prominent upregulation in post-PV-MF/post-ET-MF samples.
Since we observed a strong activation of inflammatory pathways, we analyzed NF-κB (Supplemental Fig. 4) and TNFα pathways in more detail (Supplemental Fig. 5), with KEGG pathways representing network maps of molecular interactions. Within canonical and noncanonical NF-κB pathways, BTK and IKKβ were upregulated, as well as several pro-survival target genes such as Bcl-XL, c-IAP1/2, and c-FLIP, or the inflammatory targets COX2, MIP-1β, and VCAM1. TNF, TNF receptors (TNFR) 1 and 2, and genes associated with leukocyte recruitment, inflammatory cytokines and mediators, and cell adhesion were also upregulated.
To identify deregulated pathways, we examined whether the different subtypes showed differences in gene ontology (GO) analysis compared with HCs. We observed the induction of GOs associated with inflammation such as neutrophil activation, leukocyte migration, acute inflammatory response, and others in ET, PMF, and SMF (Fig. 3, Supplemental Table 5). Surprisingly, in PV, inflammatory responses and leukocyte migration were not as upregulated as in ET. Instead, blood vessel–associated biological processes represented the strongest enriched ontologies in PV. Furthermore, RNA splicing–associated biological processes were downregulated in PMF, but not in ET, PV, and SMF, fitting the high frequency of mutations in RNA splicing–associated genes in PMF [24]. In our study, mutations in SRSF2 and SF3B1 in two PMF patients each, but not the other subtypes, were identified by NGS. Furthermore, several genes involved in the cellular response to oxidative stress and NRF2 target genes were upregulated in our investigation (Supplemental Fig. 6). NRF2 is the master regulator of the oxidative stress response by regulating the expression of a variety of antioxidative enzymes [25].
Gene ontology (GO) analysis in MPN patient- vs. HC-derived CD34+ cells. GO terms analysis of biological processes (green), and cellular components (yellow), and molecular functions (red). Graphs show the 10 most significantly enriched ontologies with over-representation of GO terms in up- (left) or downregulated (right) genes vs. HC, ranked by adj. p-value. GO terms were restricted to terms with less than 500 genes. A list of all deregulated GO terms including gene numbers and p-values is provided in Supplemental Table 5. ET, essential thrombocythemia; HC, healthy controls; PMF, primary myelofibrosis; PV, polycythemia vera; SMF, secondary myelofibrosis
The direct interaction of the clonal with normal HSPCs or endothelial and mesenchymal stromal cells plays a central role in inflammation, thrombosis, and extramedullary hematopoiesis [26]. Therefore, we investigated the expression of cell adhesion molecules and observed an upregulation of ITGB2, ITGB7, SELP, and SELPLG in all subtypes. They are involved in leukocyte adhesion and have not been investigated in MPN so far and stand as interesting targets for future studies (Supplemental Fig. 7).
Differentially regulated potential marker genes in CD34+ HSPCs from MPN subtypes
At the single gene level, we have identified the top 20 up- and downregulated genes for each subtype (Fig. 4). AREG was prominently downregulated in all entities compared to HCs. AREG encodes amphiregulin, a ligand of the epidermal growth factor receptor (EGFR). AREG binding to the EGFR activates major intracellular signaling cascades governing cell survival, proliferation, and motility [27]. On the other hand, it inhibits the growth of certain aggressive carcinoma cell lines and has an immunosuppressive function by facilitating the suppressor capacity of Tregs [28]. AREG expression was observed to be downregulated in myeloid and lymphoid neoplasms [29].
DNTT was the strongest downregulated gene in PMF and inversely correlated with driver mutation AB in CD34+ of all MPN subtypes combined (Supplemental Fig. 8). DNTT, which was also found to be downregulated in PV and SMF in our study, is expressed in pre-B and pre-T lymphocytes during early differentiation. It is a diagnostic and classification marker in ALL and low expression correlates with inferior survival [30, 31].
In addition to these downregulated genes, we also identified the upregulation of TIMD4, VCAM1, and CYBB/NOX2 in all subtypes. Furthermore, we observed increased expression of several S100 genes in CD34+ cells of one or more MPN subtypes (Fig. 5A). S100A4, S100A8, S100A9, and S100A12 are calcium- and zinc-binding proteins that function in the regulation of inflammatory processes and have been found to be associated with myelopoiesis [32]. Increased levels of S100 proteins in MPN patient-derived granulocytes and plasma as well as in PV CD34+ cells (S100A4, S100A9) have previously been reported [33]. In our GEA, we observed strongly elevated levels of S100A8, S100A9, and S100A12 in PMF, but also in ET and PV (S100A8, S100A12). S100A8/9 dimers were reported to promote tumor cell proliferation by creating a pro-inflammatory microenvironment and contribute to disease progression in MPN, whereas tasquinimod, an S100A8/S100A9 inhibitor, inhibited the MPN phenotype [34,35,36].
Expression of potential marker genes in CD34+ cells compared with unfractionated BM cells. A Gene expression levels of S100A4, S100A8, S100A9, S100A10, S100A12, TIMD4, VCAM1, CYBB, DNTT, and AREG in ET, PV, and MF patient- and HC-derived CD34+ cells (top; adj. p) or unfractionated BMMCs analyzed by RT-qPCR (bottom; n = 5 respectively; Mann–Whitney U test). B Top 10 genes that were differentially regulated in SMF vs. HC, but not the other comparisons, ranked by FC and filtered by adj. p < .05. BM, bone marrow; ET, essential thrombocythemia; FC, fold change; HC, healthy controls; MNCs, mononuclear cells; PMF, primary myelofibrosis; PV, polycythemia vera; SMF, secondary myelofibrosis. * p < .05, ** p < .01, *** p < .001, **** p < .0001 compared to HC
Deregulation of potential marker genes is restricted to the CD34+ compartment
As we have observed deregulated expression of the potential marker genes S100A4, S100A8, S100A9, S100A10, S100A12, TIMD4, VCAM1, CYBB, DNTT, and AREG, we aimed to assess whether this effect is restricted to the CD34+ compartment or also observed in unfractionated BM cells. Therefore, we performed RT-qPCR analysis of a different set of de novo and untreated JAK2V617F-positive BMMCs derived from ET, PV, and PMF patients and CD45-enriched HC cells that were isolated from femoral heads during hip replacement (n = 5).
In the case of S100A4, S100A8, S100A9, and S100A10, unfractionated BM and purified CD34+ cells showed some similarities (Fig. 5A). Strikingly, CD34+ purified cells from MPN patients showed significant upregulation of S100A12, whereas unfractionated BM showed downregulation. In addition to that, the strongly upregulated genes TIMD4, VCAM1, and CYBB only showed significant upregulation in the CD34 purified compartment. In the case of DNTT, the significant downregulation was only detected in CD34+ cells from PV and MF patients while no changes were observed in ET or in unfractionated BM cells. Similarly, AREG showed strong downregulation only in the CD34+ fraction of all MPN subtypes and unfractionated BM cells from PMF patients. These results confirm the importance of investigating the CD34+ fraction and indicate that TIMD4, VCAM1, CYBB, DNTT, AREG, and S100A12 may play a central role in MPN pathophysiology.
With the aim to identify marker genes for the progression from ET or PV to SMF, we focused on genes exclusively deregulated in SMF (Fig. 5B, Supplemental Table 4). The top 5 upregulated genes were CXCL8, CLEC1B, CMTM5, NMU, and MTSS1. Elevated blood plasma levels of the pro-inflammatory cytokine CXCL8, also termed interleukin 8 (IL-8), have been identified as a negative prognostic indicator in PMF [37]. CLEC1B is expressed by myeloid cells and modulates the secretion of cytokines [38]. The neuropeptide encoded by NMU plays a role in inflammatory diseases and its overexpression was identified in solid tumors [39]. MTSS1 and CMTM5 are tumor suppressor genes [40,41,42], although in some cancer types, MTSS1 was identified as a tumor enhancer [43, 44]. To our knowledge, an implication of CLEC1B, NMU, MTSS1, and CMTM5 in Ph− MPN is yet to be described. CXorf57, DACH1, SPTBN1, CXCR4, and FLT3 were identified as the most strongly downregulated genes. CXorf57 or RADX encodes a DNA-binding protein that prevents replication fork collapse by antagonizing the accumulation of RAD51 [45]. DACH1 was identified as a suppressor of the progression of various neoplasms and its downregulation is associated with a poor prognosis [46]. Reduced SPTBN1 levels were discovered in colon and pancreatic cancer, potentially contributing to tumor initiation or progression [47]. We are the first group to demonstrate the downregulation of RADX, DACH1, and SPTBN1 in Ph− MPN. Reduced expression of CXCR4 in CD34+ cells was recently identified as a potential diagnostic and prognostic biomarker in PMF [48]. Correspondingly, we observed strongly decreased CXCR4 expression in PMF (− 1.36 fold) and SMF (− 2.12 fold). Surprisingly, FLT3 was the most downregulated gene, although it plays an important role in the survival and proliferation of HSPCs and contributes to MF [49].
Discussion
Although the identification of the driver mutations of JAK2, CALR, and MPL found in the majority of MPN (excluding triple-negative cases) has provided substantial insight into the pathogenesis of Ph− MPN, one of the highest unmet clinical needs is the paucity of biomarkers that reliably predict the disease course. The GEA performed within this study aimed to identify differential transcriptional signatures between the MPN entities to provide novel pathogenic insights and new diagnostic or prognostic information. Within Ph− MPN, myelofibrosis (PMF or SMF) differs from ET and PV, both clinically and molecularly, with a higher prevalence of additional mutations and inflammatory changes [5, 50].
In our approach to analyze gene expression in CD34+ cells derived from ET, PV, PMF, and SMF patients, we revealed patterns that are clearly distinguishable from those of HCs. Yet, ET samples clustered closest to HCs, underlining their clinically indolent behavior (and the overall almost unimpaired life expectancy of affected patients). The discrimination of the MPN samples was not dependent on the driver mutation, which is underlined by our finding that CALR-, MPL-mutated, or triple-negative patients did not differ in the PCA or heatmaps from JAK2-mutated samples. Within the different MPN subtypes, PV patients showed gene expression patterns like those from ET patients, but overlapping patterns with PMF patients were also identified. In addition, the gene expression patterns of PMF and SMF patients revealed a high degree of similarity, corroborating the paradigm of “early” and “late” stage MPN.
Oncogenic pathway activity analysis using PROGENγ revealed that TNFα signaling and its effector pathway, NF-κB, were upregulated in all MPN subtypes (in line with findings from other groups [17, 23, 51]), but particularly in SMF, suggesting that SMF patients are characterized by a more pronounced inflammatory condition. BET bromodomain inhibitors act, among others, by inhibiting NF-κB signaling and have already shown their potential in increasing therapeutic efficacy in MPN and might be particularly effective in SMF [52]. Surprisingly, MAPK and PI3K signaling was downregulated compared to HCs. This might be explained by a CD34+-dependent activation of different MAPK (ERK, p38, JNK) or PI3K (classes IA and IB) pathways compared to the datasets that were used for training of the PROGENy algorithms. Estrogen signaling that was significantly downregulated regulates HSPC survival, proliferation, cytokine-production, and self-renewal [53]. Activation of the estrogen receptor with tamoxifen induced apoptosis in JAK2V617F HSPCs [54]. Therefore, estrogen signaling might be downregulated as a pro-survival mechanism.
Furthermore, immune system–related and pro-inflammatory pathways were upregulated in all subtypes, particularly in PMF and SMF. JAK1/2- and STAT3/5-mediated inflammatory processes play an important role in the pathogenesis and progression of MPN [55]. Among the key players in MPN pathophysiology are clonal HSPCs, as well as inflammatory cytokines and their interplay with endothelial and mesenchymal stromal cells (MSCs) [56]. The pro-inflammatory signatures are corroborated by the activation of TNF and NF-κB signaling pathways, identified in our PROGENγ analysis, and the effectiveness of inflammation modulating drugs such as interferon-alpha (IFNa).
We further identified the induction of GO terms associated with inflammation in all four MPN entities. These ontologies were most significantly enriched in PMF and SMF. In contrast, only PV had significantly downregulated ontologies comprising RNA splicing pathways. Mutations in RNA splicing–associated genes such as SRSF2, SF3B1, and U2AF1 are often observed in MPN suggesting a pathophysiological role [5]. SRSF2 mutations are associated with poor prognosis in PMF and enriched in leukemic transformation. These data support the poor risk profile of SRSF2 mutation in PV and ET patients.
In addition, we observed enrichment of oxidative stress-associated genes in all four entities. The aberrant activation of JAK2 induces DNA damage by cell cycle–induced DNA damage in the hematopoietic stem cell compartment [57]. The frequency of these damaging events that lead to genomic instability is further increased by the accumulation of reactive oxygen species (ROS) [58]. ROS can stabilize HIF-1α and induce aberrant HIF signaling accompanied by enhanced glycolysis that both were identified as potential targets in MPN [59, 60]. On the other hand, excessive ROS levels induce apoptosis leading to the activation of NRF2 as a regulator of the cellular resistance to oxidative stress in CD34+ MPN cells. In contrast, downregulation of NRF2 signaling was reported in MPN patient-derived whole blood cells fueling the expansion of the hematopoietic progenitor pool [61]. We hypothesize a different role of NRF2 in CD34+ MPN cells as NRF2 is involved in the regulation of HSPC function by increasing quiescence and self-renewal [62].
A set of deregulated genes was identified that, beyond their putative role in MPN pathogenesis, may also have prognostic and therapeutic significance. AREG was downregulated in the CD34+ fraction of all four MPN entities and unfractionated BM from PMF patients. Although its molecular function in the context of MPN remains to be elucidated, it might serve as a molecular disease marker. The downregulation of DNTT, which is implicated in B cell development, might be indicative of the shift of HSPC fate towards the myeloid lineage.
We also identified several upregulated genes that might contribute to the pathogenesis of MPN such as the pro-inflammatory alarmins S100A8 and S100A9. Both proteins are TLR4 ligands that activate NF-κB and induce the secretion of pro-inflammatory cytokines [63]. In MPN, the S100A8/A9-induced erythroid differentiation block might contribute to the high risk of leukemic transformation in PMF and SMF, whereas its expression is considerably lower in ET and PV [64]. Strikingly, the S100A8/S100A9 inhibitor tasquinimod significantly ameliorated the MPN phenotype and fibrosis in a JAK2V617F mouse model [35]. Furthermore, increased S100A8/S100A9 expression seems to play a central role in the telomere-associated inflammatory environment found in CML [65]. CYBB/NOX2 that was upregulated in all subtypes is a super-oxide-generating enzyme that interacts with S100A8/9 and might contribute to elevated ROS levels in MPN patients promoting the acquisition of additional mutations and the stabilization of HIF-1α [60, 66]. Elevated levels of S100A10 were reported to attenuate the pro-apoptotic effects of BAD [67]. Furthermore, it contributes to coagulopathy in AML by increasing the affinity of tPa for plasminogen leading to hyperfibrinolysis, which is associated with bleeding [68]. S100A12 is expressed in several tumors mediating proliferation, invasion, and migration [69].
TIMD4 that was upregulated in all subtypes was observed to induce T cell exhaustion in cancer and promote colorectal cancer by activating angiogenesis and recruitment of tumor-associated macrophages [70, 71]. In PMF, TIMD4 was reported to recruit monocytes/macrophages to the BM [72]. VCAM1, which was also elevated in our GEA, contributes to the clinical phenomena of thrombosis and extramedullary hematopoiesis occurring in MPN patients [73].
RT-qPCR analysis of these genes in unfractionated BM-derived cells revealed that their differential regulation is mainly restricted to CD34+ cells underlining the potential implication of these marker genes specifically within more primitive populations. This was particularly prominent for S100A12 that was strongly upregulated in the CD34+ compartment but downregulated in unfractionated BM in all subtypes. We also identified potential marker genes for a transition from ET or PV to SMF (such as CLEC1B, NMU, or DACH1) that were strongly deregulated only in SMF. Within the top 10 up- and downregulated genes, we also observed elevated expression of MTSS1 that is suppressed in high-risk AML but increased in patients with better clinical outcome [41].
Nevertheless, limitations of our study need to be discussed as well. The sample size of the SMF group is relatively small and all-female. The mutational landscape in the ET group is heterogeneous combining JAK2-, CALR-, MPL-mutated, and triple-negative patients possibly leading to a bias in the gene expression patterns between the different subtypes.
In summary, our investigation has demonstrated that CD34+ cells derived from PMF and SMF patients differ from ET and PV patients in the expression of single genes and the activation of signaling pathways. The different MPN subtypes are based on the mutual activation of the JAK-STAT pathway. Although this explains similarities such as the 47 jointly deregulated genes and a general activation of inflammatory signaling pathways, we observed differences between the subtypes that could contribute to modeling the disease transition from early to late stage MPN. Furthermore, several of these genes and pathways might constitute potential disease markers or candidates for targeted therapy, especially those genes deregulated only in the CD34+ fraction. Noteworthy is the strong activation of inflammatory pathways confirming that anti-inflammatory drugs such as ruxolitinib or BET bromodomain protein inhibitors or disease-modulating agents such as IFNa constitute a promising approach in Ph− MPN, especially in later stages.
Availability of data and material
Microarray data have been deposited at GEO under accession number GSE174060.
Code availability
Not applicable.
References
Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, Chen D, Vladimer GI, Bagienski K, Milanesi C, Casetti IC, Sant’Antonio E, Ferretti V, Elena C, Schischlik F, Cleary C, Six M, Schalling M, Schonegger A, Bock C, Malcovati L, Pascutto C, Superti-Furga G, Cazzola M, Kralovics R, (2013) Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 369(25):2379–2390. https://doi.org/10.1056/NEJMoa1311347
Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC (2005) A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352(17):1779–1790. https://doi.org/10.1056/NEJMoa051113
Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD, Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP, Teague JW, O’Meara S, McLaren S, Bianchi M, Silber Y, Dimitropoulou D, Bloxham D, Mudie L, Maddison M, Robinson B, Keohane C, Maclean C, Hill K, Orchard K, Tauro S, Du MQ, Greaves M, Bowen D, Huntly BJP, Harrison CN, Cross NCP, Ron D, Vannucchi AM, Papaemmanuil E, Campbell PJ, Green AR (2013) Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 369(25):2391–2405. https://doi.org/10.1056/NEJMoa1312542
Tefferi A (2010) Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 24(6):1128–1138. https://doi.org/10.1038/leu.2010.69
Vainchenker W, Kralovics R (2017) Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 129(6):667–679. https://doi.org/10.1182/blood-2016-10-695940
Guglielmelli P, Lasho TL, Rotunno G, Mudireddy M, Mannarelli C, Nicolosi M, Pacilli A, Pardanani A, Rumi E, Rosti V, Hanson CA, Mannelli F, Ketterling RP, Gangat N, Rambaldi A, Passamonti F, Barosi G, Barbui T, Cazzola M, Vannucchi AM, Tefferi A (2018) MIPSS70: mutation-enhanced international prognostic score system for transplantation-age patients with primary myelofibrosis. J Clin Oncol 36(4):310–318. https://doi.org/10.1200/JCO.2017.76.4886
Grinfeld J, Nangalia J, Baxter EJ, Wedge DC, Angelopoulos N, Cantrill R, Godfrey AL, Papaemmanuil E, Gundem G, MacLean C, Cook J, O’Neil L, O’Meara S, Teague JW, Butler AP, Massie CE, Williams N, Nice FL, Andersen CL, Hasselbalch HC, Guglielmelli P, McMullin MF, Vannucchi AM, Harrison CN, Gerstung M, Green AR, Campbell PJ (2018) Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med 379(15):1416–1430. https://doi.org/10.1056/NEJMoa1716614
Hasselbalch HC (2019) Time for revival of the red blood cell count and red cell mass in the differential diagnosis between essential thrombocythemia and polycythemia vera? Haematologica 104(11):2119–2125. https://doi.org/10.3324/haematol.2019.229039
Pardanani A, Vannucchi AM, Passamonti F, Cervantes F, Barbui T, Tefferi A (2011) JAK inhibitor therapy for myelofibrosis: critical assessment of value and limitations. Leukemia 25(2):218–225. https://doi.org/10.1038/leu.2010.269
Vannucchi AM, Harrison CN (2017) Emerging treatments for classical myeloproliferative neoplasms. Blood 129(6):693–703. https://doi.org/10.1182/blood-2016-10-695965
Patel AA (2020) Odenike O (2020) Genomics of MPN progression. Hematology Am Soc Hematol Educ Program 1:440–449. https://doi.org/10.1182/hematology.2020000129
Berkofsky-Fessler W, Buzzai M, Kim MK, Fruchtman S, Najfeld V, Min DJ, Costa FF, Bischof JM, Soares MB, McConnell MJ, Zhang W, Levine R, Gilliland DG, Calogero R, Licht JD (2010) Transcriptional profiling of polycythemia vera identifies gene expression patterns both dependent and independent from the action of JAK2V617F. Clin Cancer Res 16(17):4339–4352. https://doi.org/10.1158/1078-0432.CCR-10-1092
Catani L, Zini R, Sollazzo D, Ottaviani E, Vannucchi AM, Ferrari S, Baccarani M, Vianelli N, Lemoli RM, Manfredini R (2009) Molecular profile of CD34+ stem/progenitor cells according to JAK2V617F mutation status in essential thrombocythemia. Leukemia 23(5):997–1000. https://doi.org/10.1038/leu.2008.357
Guglielmelli P, Zini R, Bogani C, Salati S, Pancrazzi A, Bianchi E, Mannelli F, Ferrari S, Le Bousse-Kerdiles MC, Bosi A, Barosi G, Migliaccio AR, Manfredini R, Vannucchi AM (2007) Molecular profiling of CD34+ cells in idiopathic myelofibrosis identifies a set of disease-associated genes and reveals the clinical significance of Wilms’ tumor gene 1 (WT1). Stem Cells 25(1):165–173. https://doi.org/10.1634/stemcells.2006-0351
Čokić VP, Mossuz P, Han J, Socoro N, Beleslin-Čokić BB, Mitrović O, Subotički T, Diklić M, Leković D, Gotič M, Puri RK, Noguchi CT, Schechter AN (2015) Microarray and proteomic analyses of myeloproliferative neoplasms with a highlight on the mTOR signaling pathway. PLoS ONE 10(8):1–23. https://doi.org/10.1371/journal.pone.0135463
Psaila B, Wang G, Rodriguez-Meira A, Li R, Heuston EF, Murphy L, Yee D, Hitchcock IS, Sousos N, O’Sullivan J, Anderson S, Senis YA, Weinberg OK, Calicchio ML, Center NIHIS, Iskander D, Royston D, Milojkovic D, Roberts I, Bodine DM, Thongjuea S, Mead AJ (2020) Single-cell analyses reveal megakaryocyte-biased hematopoiesis in myelofibrosis and identify mutant clone-specific targets. Mol Cell 78 (3):477–492 e478. doi:https://doi.org/10.1016/j.molcel.2020.04.008
Nam AS, Kim KT, Chaligne R, Izzo F, Ang C, Taylor J, Myers RM, Abu-Zeinah G, Brand R, Omans ND, Alonso A, Sheridan C, Mariani M, Dai X, Harrington E, Pastore A, Cubillos-Ruiz JR, Tam W, Hoffman R, Rabadan R, Scandura JM, Abdel-Wahab O, Smibert P, Landau DA (2019) Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 571(7765):355–360. https://doi.org/10.1038/s41586-019-1367-0
Kirschner M, Maurer A, Wlodarski MW, Ventura Ferreira MS, Bouillon AS, Halfmeyer I, Blau W, Kreuter M, Rosewich M, Corbacioglu S, Beck J, Schwarz M, Bittenbring J, Radsak MP, Wilk CM, Koschmieder S, Begemann M, Kurth I, Schemionek M, Brummendorf TH, Beier F (2018) Recurrent somatic mutations are rare in patients with cryptic dyskeratosis congenita. Leukemia 32(8):1762–1767. https://doi.org/10.1038/s41375-018-0125-x
Czech J, Cordua S, Weinbergerova B, Baumeister J, Crepcia A, Han L, Maie T, Costa IG, Denecke B, Maurer A, Schubert C, Feldberg K, Gezer D, Brummendorf TH, Muller-Newen G, Mayer J, Racil Z, Kubesova B, Knudsen T, Sorensen AL, Holmstrom M, Kjaer L, Skov V, Larsen TS, Hasselbalch HC, Chatain N, Koschmieder S (2019) JAK2V617F but not CALR mutations confer increased molecular responses to interferon-alpha via JAK1/STAT1 activation. Leukemia 33(4):995–1010. https://doi.org/10.1038/s41375-018-0295-6
Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127. https://doi.org/10.1093/biostatistics/kxj037
Nangalia J, Green AR (2017) Myeloproliferative neoplasms: from origins to outcomes. Blood 130 (23):blood-2017–2006–782037. doi:https://doi.org/10.1182/blood-2017-06-782037
Schubert M, Klinger B, Klunemann M, Sieber A, Uhlitz F, Sauer S, Garnett MJ, Bluthgen N, Saez-Rodriguez J (2018) Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun 9(1):20. https://doi.org/10.1038/s41467-017-02391-6
Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, De Groote S, Papalexi E, Hanasoge Somasundara AV, Cordner K, Keller M, Farnoud N, Medina J, McGovern E, Reyes J, Roberts J, Witkin M, Rapaport F, Teruya-Feldstein J, Qi J, Rampal R, Bernstein BE, Bradner JE, Levine RL (2018) Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell 33 (1):29–43 e27. doi:https://doi.org/10.1016/j.ccell.2017.11.009
Hautin M, Mornet C, Chauveau A, Bernard D, Corcos L, Lippert E (2020) Splicing anomalies in myeloproliferative neoplasms: paving the way for new therapeutic venues. Cancers (Basel) 12 (8). doi:https://doi.org/10.3390/cancers12082216
Ma Q (2013) Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol 53:401–426. https://doi.org/10.1146/annurev-pharmtox-011112-140320
Chatain N, Koschmieder S, Jost E (2020) Role of inflammatory factors during disease pathogenesis and stem cell transplantation in myeloproliferative neoplasms. Cancers (Basel) 12 (8). doi:https://doi.org/10.3390/cancers12082250
Berasain C, Avila MA (2014) Amphiregulin. Semin Cell Dev Biol 28:31–41. https://doi.org/10.1016/j.semcdb.2014.01.005
Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, Treuting PM, Rudensky AY (2015) A distinct function of regulatory T cells in tissue protection. Cell 162(5):1078–1089. https://doi.org/10.1016/j.cell.2015.08.021
Haouas H, Haouas S, Uzan G, Hafsia A (2010) Identification of new markers discriminating between myeloid and lymphoid acute leukemia. Hematology 15(4):193–203. https://doi.org/10.1179/102453310X12647083620769
Huh YO, Smith TL, Collins P, Bueso-Ramos C, Albitar M, Kantarjian HM, Pierce SA, Freireich EJ (2000) Terminal deoxynucleotidyl transferase expression in acute myelogenous leukemia and myelodysplasia as determined by flow cytometry. Leuk Lymphoma 37(3–4):319–331. https://doi.org/10.3109/10428190009089432
Paiva AS, Paiva HDDO, Cavalcanti GB Jr, Bahia F, Freitas RV, Silveira LS Jr, Silva LK, Vasconcelos RC, Silva DG, Vl S, Gil EA, Januario G, Fernandes M, Jardim AS (2018) Importance of detection of intracellular myeloperoxidase, CD13, CD79a, CD22, CD3 and terminal deoxynucleotidyl transferase by flow cytometry diagnosis of acute leukemias. Blood 132(Supplement 1):5194–5194. https://doi.org/10.1182/blood-2018-99-118880
Goyette J, Geczy CL (2011) Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids 41(4):821–842. https://doi.org/10.1007/s00726-010-0528-0
Kovacic M, Mitrovic-Ajtic O, Beleslin-Cokic B, Djikic D, Suboticki T, Diklic M, Lekovic D, Gotic M, Mossuz P, Cokic VP (2018) TLR4 and RAGE conversely mediate pro-inflammatory S100A8/9-mediated inhibition of proliferation-linked signaling in myeloproliferative neoplasms. Cell Oncol (Dordr) 41(5):541–553. https://doi.org/10.1007/s13402-018-0392-6
Turovskaya O, Foell D, Sinha P, Vogl T, Newlin R, Nayak J, Nguyen M, Olsson A, Nawroth PP, Bierhaus A, Varki N, Kronenberg M, Freeze HH, Srikrishna G (2008) RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis 29(10):2035–2043. https://doi.org/10.1093/carcin/bgn188
Leimkuhler NB, Gleitz HFE, Ronghui L, Snoeren IAM, Fuchs SNR, Nagai JS, Banjanin B, Lam KH, Vogl T, Kuppe C, Stalmann USA, Busche G, Kreipe H, Gutgemann I, Krebs P, Banz Y, Boor P, Tai EW, Brummendorf TH, Koschmieder S, Crysandt M, Bindels E, Kramann R, Costa IG, Schneider RK (2020) Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell. https://doi.org/10.1016/j.stem.2020.11.004
Gleitz HFE, Dugourd AJF, Leimkuhler NB, Snoeren IAM, Fuchs SNR, Menzel S, Ziegler S, Kroger N, Triviai I, Busche G, Kreipe H, Banjanin B, Pritchard JE, Hoogenboezem R, Bindels EM, Schumacher N, Rose-John S, Elf S, Saez-Rodriguez J, Kramann R, Schneider RK (2020) Increased CXCL4 expression in hematopoietic cells links inflammation and progression of bone marrow fibrosis in MPN. Blood 136(18):2051–2064. https://doi.org/10.1182/blood.2019004095
Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A (2011) Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol 29(10):1356–1363. https://doi.org/10.1200/JCO.2010.32.9490
Colonna M, Samaridis J, Angman L (2000) Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. Eur J Immunol 30(2):697–704. https://doi.org/10.1002/1521-4141(200002)30:2%3c697::AID-IMMU697%3e3.0.CO;2-M
Moriyama M, Sato T, Inoue H, Fukuyama S, Teranishi H, Kangawa K, Kano T, Yoshimura A, Kojima M (2005) The neuropeptide neuromedin U promotes inflammation by direct activation of mast cells. J Exp Med 202(2):217–224. https://doi.org/10.1084/jem.20050248
Schemionek M, Herrmann O, Reher MM, Chatain N, Schubert C, Costa IG, Hanzelmann S, Gusmao EG, Kintsler S, Braunschweig T, Hamilton A, Helgason GV, Copland M, Schwab A, Muller-Tidow C, Li S, Holyoake TL, Brummendorf TH, Koschmieder S (2016) Mtss1 is a critical epigenetically regulated tumor suppressor in CML. Leukemia 30(4):823–832. https://doi.org/10.1038/leu.2015.329
Schemionek M, Kharabi Masouleh B, Klaile Y, Krug U, Hebestreit K, Schubert C, Dugas M, Buchner T, Wormann B, Hiddemann W, Berdel WE, Brummendorf TH, Muller-Tidow C, Koschmieder S (2015) Identification of the adapter molecule MTSS1 as a potential oncogene-specific tumor suppressor in acute myeloid leukemia. PLoS ONE 10(5):e0125783. https://doi.org/10.1371/journal.pone.0125783
Shao L, Cui Y, Li H, Liu Y, Zhao H, Wang Y, Zhang Y, Ng KM, Han W, Ma D, Tao Q (2007) CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res 13(19):5756–5762. https://doi.org/10.1158/1078-0432.CCR-06-3082
Wang D, Xu MR, Wang T, Li T, Zhu J (2011) MTSS1 overexpression correlates with poor prognosis in colorectal cancer. J Gastrointest Surg 15(7):1205–1212. https://doi.org/10.1007/s11605-011-1546-2
Mertz KD, Pathria G, Wagner C, Saarikangas J, Sboner A, Romanov J, Gschaider M, Lenz F, Neumann F, Schreiner W, Nemethova M, Glassmann A, Lappalainen P, Stingl G, Small JV, Fink D, Chin L, Wagner SN (2014) MTSS1 is a metastasis driver in a subset of human melanomas. Nat Commun 5:3465. https://doi.org/10.1038/ncomms4465
Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, Sharan SK, Wessel SR, Sathe AA, Zhao R, Cortez D (2017) RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol Cell 67 (3):374–386 e375. doi:https://doi.org/10.1016/j.molcel.2017.06.023
Liu Y, Zhou R, Yuan X, Han N, Zhou S, Xu H, Guo M, Yu S, Zhang C, Yin T, Wu K (2015) DACH1 is a novel predictive and prognostic biomarker in hepatocellular carcinoma as a negative regulator of Wnt/beta-catenin signaling. Oncotarget 6(11):8621–8634. https://doi.org/10.18632/oncotarget.3281
Chen S, Li J, Zhou P, Zhi X (2020) SPTBN1 and cancer, which links? J Cell Physiol 235(1):17–25. https://doi.org/10.1002/jcp.28975
Barosi G, Rosti V, Catarsi P, Villani L, Abba C, Carolei A, Magrini U, Gale RP, Massa M, Campanelli R (2020) Reduced CXCR4-expression on CD34-positive blood cells predicts outcomes of persons with primary myelofibrosis. Leukemia. https://doi.org/10.1038/s41375-020-0926-6
Verstovsek S, Komrokji RS (2015) A comprehensive review of pacritinib in myelofibrosis. Future Oncol 11(20):2819–2830. https://doi.org/10.2217/fon.15.200
Hasselbalch HC, Bjorn ME (2015) MPNs as inflammatory diseases: the evidence, consequences, and perspectives. Mediators Inflamm 2015:102476. https://doi.org/10.1155/2015/102476
Fisher DAC, Malkova O, Engle EK, Miner CA, Fulbright MC, Behbehani GK, Collins TB, Bandyopadhyay S, Zhou A, Nolan GP, Oh ST (2017) Mass cytometry analysis reveals hyperactive NF Kappa B signaling in myelofibrosis and secondary acute myeloid leukemia. Leukemia 31(9):1962–1974. https://doi.org/10.1038/leu.2016.377
Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, De Groote S, Papalexi E, Hanasoge Somasundara AV, Cordner K, Keller M, Farnoud N, Medina J, McGovern E, Reyes J, Roberts J, Witkin M, Rapaport F, Teruya-Feldstein J, Qi J, Rampal R, Bernstein BE, Bradner JE, Levine RL (2018) Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell 33(4):785–787. https://doi.org/10.1016/j.ccell.2018.03.024
Sanchez-Aguilera A, Arranz L, Martin-Perez D, Garcia-Garcia A, Stavropoulou V, Kubovcakova L, Isern J, Martin-Salamanca S, Langa X, Skoda RC, Schwaller J, Mendez-Ferrer S (2014) Estrogen signaling selectively induces apoptosis of hematopoietic progenitors and myeloid neoplasms without harming steady-state hematopoiesis. Cell Stem Cell 15(6):791–804. https://doi.org/10.1016/j.stem.2014.11.002
Harrison C, Baxter J, Boucher RH, McKerrell T, Jackson A, Fletcher RS, Mead AJ, Knapper S, Ewing J, Butt NM, Jain M, Francis S, Clark FJ, Coppell J, McMullin MF, Wadelin F, Narayanan S, Milojkovic D, Drummond MW, Sekhar M, Paramor M, Roberts J, Glover R, Harris E, Grassi L, Rodriguez-Meira A, Carroll J, Martín J-E, Corbizi Fattori G, Fang Z, Godfrey AL, Méndez-Ferrer S (2020) Effects of tamoxifen on the mutant allele burden and disease course in patients with myeloproliferative neoplasms - results of the Tamarin Study. Blood 136(Supplement 1):33–35. https://doi.org/10.1182/blood-2020-134764
Koschmieder S, Mughal TI, Hasselbalch HC, Barosi G, Valent P, Kiladjian JJ, Jeryczynski G, Gisslinger H, Jutzi JS, Pahl HL, Hehlmann R, Maria Vannucchi A, Cervantes F, Silver RT, Barbui T (2016) Myeloproliferative neoplasms and inflammation: whether to target the malignant clone or the inflammatory process or both. Leukemia 30(5):1018–1024. https://doi.org/10.1038/leu.2016.12
Koschmieder S, Chatain N (2020) Role of inflammation in the biology of myeloproliferative neoplasms. Blood Rev 42:100711. https://doi.org/10.1016/j.blre.2020.100711
Nieborowska-Skorska M, Maifrede S, Dasgupta Y, Sullivan K, Flis S, Le BV, Solecka M, Belyaeva EA, Kubovcakova L, Nawrocki M, Kirschner M, Zhao H, Prchal JT, Piwocka K, Moliterno AR, Wasik M, Koschmieder S, Green TR, Skoda RC, Skorski T (2017) Ruxolitinib-induced defects in DNA repair cause sensitivity to PARP inhibitors in myeloproliferative neoplasms. Blood 130(26):2848–2859. https://doi.org/10.1182/blood-2017-05-784942
Marty C, Lacout C, Droin N, Le Couédic JP, Ribrag V, Solary E, Vainchenker W, Villeval JL, Plo I (2013) A role for reactive oxygen species in JAK2 V617F myeloproliferative neoplasm progression. Leukemia 27(11):2187–2195. https://doi.org/10.1038/leu.2013.102
Nageswara Rao T, Hansen N, Hilfiker J, Rai S, Majewska JM, Lekovic D, Gezer D, Andina N, Galli S, Cassel T, Geier F, Delezie J, Nienhold R, Hao-Shen H, Beisel C, Di Palma S, Dimeloe S, Trebicka J, Wolf D, Gassmann M, Fan TW, Lane AN, Handschin C, Dirnhofer S, Kroger N, Hess C, Radimerski T, Koschmieder S, Cokic VP, Skoda RC (2019) JAK2 mutant hematopoietic cells display metabolic alterations that can be targeted to treat myeloproliferative neoplasms. Blood. https://doi.org/10.1182/blood.2019000162
Baumeister J, Chatain N, Hubrich A, Maie T, Costa IG, Denecke B, Han L, Kustermann C, Sontag S, Sere K, Strathmann K, Zenke M, Schuppert A, Brummendorf TH, Kranc KR, Koschmieder S, Gezer D (2019) Hypoxia-inducible factor 1 (HIF-1) is a new therapeutic target in JAK2V617F-positive myeloproliferative neoplasms. Leukemia. https://doi.org/10.1038/s41375-019-0629-z
Hasselbalch HC, Thomassen M, Riley CH, Kjaer L, Larsen TS, Jensen MK, Bjerrum OW, Kruse TA, Skov V (2014) Whole blood transcriptional profiling reveals deregulation of oxidative and antioxidative defence genes in myelofibrosis and related neoplasms. Potential implications of downregulation of Nrf2 for genomic instability and disease progression. PLoS One 9 (11):e112786. doi:https://doi.org/10.1371/journal.pone.0112786
Tsai JJ, Dudakov JA, Takahashi K, Shieh JH, Velardi E, Holland AM, Singer NV, West ML, Smith OM, Young LF, Shono Y, Ghosh A, Hanash AM, Tran HT, Moore MA, van den Brink MR (2013) Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol 15(3):309–316. https://doi.org/10.1038/ncb2699
Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J (2009) The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol 86(3):557–566. https://doi.org/10.1189/jlb.1008647
Abdulkarim K, Girodon F, Johansson P, Maynadie M, Kutti J, Carli PM, Bovet E, Andreasson B (2009) AML transformation in 56 patients with Ph- MPD in two well defined populations. Eur J Haematol 82(2):106–111. https://doi.org/10.1111/j.1600-0609.2008.01163.x
Braig M, Pallmann N, Preukschas M, Steinemann D, Hofmann W, Gompf A, Streichert T, Braunschweig T, Copland M, Rudolph KL, Bokemeyer C, Koschmieder S, Schuppert A, Balabanov S, Brummendorf TH (2014) A ‘telomere-associated secretory phenotype’ cooperates with BCR-ABL to drive malignant proliferation of leukemic cells. Leukemia 28(10):2028–2039. https://doi.org/10.1038/leu.2014.95
Chan SL, Baumbach GL (2013) Deficiency of Nox2 prevents angiotensin II-induced inward remodeling in cerebral arterioles. Front Physiol 4:133. https://doi.org/10.3389/fphys.2013.00133
Hsu SY, Kaipia A, Zhu L, Hsueh AJ (1997) Interference of BAD (Bcl-xL/Bcl-2-associated death promoter)-induced apoptosis in mammalian cells by 14–3-3 isoforms and P11. Mol Endocrinol 11(12):1858–1867. https://doi.org/10.1210/mend.11.12.0023
Brenner AK, Bruserud O (2018) S100 proteins in acute myeloid leukemia. Neoplasia 20(12):1175–1186. https://doi.org/10.1016/j.neo.2018.09.007
Rouleau P, Vandal K, Ryckman C, Poubelle PE, Boivin A, Talbot M, Tessier PA (2003) The calcium-binding protein S100A12 induces neutrophil adhesion, migration, and release from bone marrow in mouse at concentrations similar to those found in human inflammatory arthritis. Clin Immunol 107(1):46–54. https://doi.org/10.1016/s1521-6616(02)00043-8
Cheng L, Ruan Z (2015) Tim-3 and Tim-4 as the potential targets for antitumor therapy. Hum Vaccin Immunother 11(10):2458–2462. https://doi.org/10.1080/21645515.2015.1056953
Tan X, Zhang Z, Yao H, Shen L (2018) Tim-4 promotes the growth of colorectal cancer by activating angiogenesis and recruiting tumor-associated macrophages via the PI3K/AKT/mTOR signaling pathway. Cancer Lett 436:119–128. https://doi.org/10.1016/j.canlet.2018.08.012
Ozono Y, Shide K, Kameda T, Kamiunten A, Tahira Y, Sekine M, Akizuki K, Nakamura K, Iwakiri H, Sueta M, Hidaka T, Kubuki Y, Yamamoto S, Hasuike S, Sawaguchi A, Nagata K, Shimoda K (2021) Neoplastic fibrocytes play an essential role in bone marrow fibrosis in Jak2V617F-induced primary myelofibrosis mice. Leukemia 35(2):454–467. https://doi.org/10.1038/s41375-020-0880-3
Edelmann B, Gupta N, Schnoeder TM, Oelschlegel AM, Shahzad K, Goldschmidt J, Philipsen L, Weinert S, Ghosh A, Saalfeld FC, Nimmagadda SC, Muller P, Braun-Dullaeus R, Mohr J, Wolleschak D, Kliche S, Amthauer H, Heidel FH, Schraven B, Isermann B, Muller AJ, Fischer T (2018) JAK2-V617F promotes venous thrombosis through beta1/beta2 integrin activation. J Clin Invest 128(10):4359–4371. https://doi.org/10.1172/JCI90312
Acknowledgements
This work was supported by a grant to TB and AS (grant O1-6) from the Interdisziplinäre Zentrum für Klinische Forschung (IZKF) Aachen of the Faculty of Medicine at Uniklinik RWTH Aachen, by a grant from the Deutsche Forschungsgemeinschaft (DFG) to SK (KO2155/6-1), and by DFG grants within the Clinical Research Unit 344 to TB (BR1782/5-1), SK (KO2155/7-1), and IC (GE2811/4-1). In addition, the work was supported by the Genomics Facility, a core facility of the IZKF Aachen at Uniklinik RWTH Aachen under the supervision of Bernd Denecke. Biomaterial samples were provided by the RWTH centralized Biomaterial Bank Aachen (RWTH cBMB, Aachen, Germany) in accordance with the regulations of the biomaterial bank and the approval of the ethics committee of the medical faculty, RWTH Aachen. The work was supported by institutional support from the Ministry of Health of Czech Republic to ZR (#00023736). Supported by MH CZ—DRO (FNBr, 65269705) to SM.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by a grant to TB and AS (grant O1-6) from the Interdisziplinäre Zentrum für Klinische Forschung (IZKF) Aachen of the Faculty of Medicine at Uniklinik RWTH Aachen, by a grant from the Deutsche Forschungsgemeinschaft (DFG) to SK (KO2155/6–1), and by DFG grants within the Clinical Research Unit 344 to TB (BR1782/5–1), SK (KO2155/7–1), and IC (GE2811/4–1). The work was supported by institutional support from the Ministry of Health of Czech Republic to ZR (#00023736). Supported by MH CZ—DRO (FNBr, 65269705) to JM.
Author information
Authors and Affiliations
Contributions
Julian Baumeister designed and performed the experiments and analyzed the data; Julian Baumeister and Deniz Gezer wrote the paper; Tiago Maié and Ivan Costa performed the bioinformatics analysis; Lin Gan performed and analyzed the gene expression arrays; Angela Maurer performed the NGS analysis and reviewed the variants; Nicolas Chatain, Marcelo August Szymanski de Toledo, Tim H. Brümmendorf, Steffen Koschmieder, Andreas Schuppert, and Deniz Gezer designed the experiments and analyzed the data; Nicolas Chatain and Steffen Koschmieder participated in writing the paper; Jörg Eschweiler, Jiri Mayer, Zdenek Racil, Blanka Kubesova, and Barbora Weinbergerova provided BM samples from JAK2V617F-positive untreated MPN patients or healthy donors and participated in writing the paper; Andreas Schuppert and Tim H. Brümmendorf evaluated the data and participated in writing the paper; and all authors read, critically revised, and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
PB and BM samples from MPN patients were obtained at the Department of Hematology, Oncology, Hemostaseology, and Stem Cell Transplantation at Uniklinik RWTH Aachen after written informed consent and approval of the local ethics committee (EK127/12). Unfractionated BM samples from JAK2V617F-positive untreated MPN patients within the CELL project — MPN database (MIND) — after written informed consent were provided from the Department of Internal Medicine, Hematology and Oncology of Brno University Hospital, Czech Republic. Femoral heads were obtained from the Department of Orthopedics from the Uniklinik RWTH Aachen, each after written informed consent and in compliance with the local ethics committee (EK300/13).
Consent to participate
See “Ethics approval.”
Consent for publication
The patients’ written informed consent includes a statement that their data will be published.
Conflict of interest
SK reports advisory board activity for Pfizer, Incyte/Ariad, Novartis, AOP Pharma, BMS, CTI, Roche, Baxalta, Sanofi; honoraria from Novartis, BMS, Pfizer, Incyte/Ariad, Shire, Roche, AOP Pharma, Janssen; research funding from Novartis Foundation, BMS, Novartis; and other financial disclosures (i.e., travel support) from Alexion, Novartis, BMS, Incyte/Ariad, AOP Pharma, Baxalta, CTI, Pfizer, Sanofi, Celgene, Shire, and Janssen. THB reports consultancy from Pfizer, Novartis, Janssen, Merck, Incyte/Ariad and research funding from Pfizer and Novartis. DG reports advisory board activity for AMGEN, Celgene, and TAKEDA; travel grants for AMGEN, Celgene, and TAKEDA; and was invited speaker for AMGEN, Celgene, BMS, and TAKEDA. The remaining authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Baumeister, J., Maié, T., Chatain, N. et al. Early and late stage MPN patients show distinct gene expression profiles in CD34+ cells. Ann Hematol 100, 2943–2956 (2021). https://doi.org/10.1007/s00277-021-04615-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-021-04615-8