Abstract
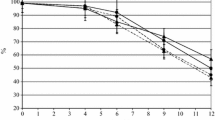
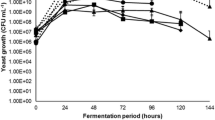
Non-Saccharomyces (NS) species that are either naturally present in grape must or added in mixed fermentation with S. cerevisiae may impact the wine’s chemical composition and sensory properties. NS yeasts are prevailing during prefermentation and early stages of alcoholic fermentation. However, obtaining the correct balance between S. cerevisiae and NS species is still a critical issue: if S. cerevisiae outcompetes the non-Saccharomyces, it may minimize their impact, while conversely if NS take over S. cerevisiae, it may result in stuck or sluggish fermentations. Here, we propose an original strategy to promote the non-Saccharomyces consortium during the prefermentation stage while securing fermentation completion: the use of a long lag phase S. cerevisiae. Various fermentations in a Sauvignon Blanc with near isogenic S. cerevisiae displaying short or long lag phase were compared. Fermentations were performed with or without a consortium of five non-Saccharomyces yeasts (Hanseniaspora uvarum, Candida zemplinina, Metschnikowia spp., Torulaspora delbrueckii, and Pichia kluyveri), mimicking the composition of natural NS community in grape must. The sensorial analysis highlighted the positive impact of the long lag phase on the wine fruitiness and complexity. Surprisingly, the presence of NS modified only marginally the wine composition but significantly impacted the lag phase of S. cerevisiae. The underlying mechanisms are still unclear, but it is the first time that a study suggests that the wine composition can be affected by the lag phase duration per se. Further experiments should address the suitability of the use of long lag phase S. cerevisiae in winemaking.





Similar content being viewed by others
References
Albergaria H, Francisco D, Gori K, Arneborg N, Gírio F (2010) Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl Microbiol Biotechnol 86:965–972. https://doi.org/10.1007/s00253-009-2409-6
Andorra I, Landi S, Mas A, Esteve-Zarzoso B, Guillamon JM (2010) Effect of fermentation temperature on microbial population evolution using culture-independent and dependent techniques. Food Res Int 43:773–779. https://doi.org/10.1016/j.foodres.2009.11.014
Andorra I, Monteiro M, Esteve-Zarzoso B, Albergaria H, Mas A (2011) Analysis and direct quantification of Saccharomyces cerevisiae and Hanseniaspora guilliermondii populations during alcoholic fermentation by fluorescence in situ hybridization, flow cytometry and quantitative PCR. Food Microbiol 28:1483–1491. https://doi.org/10.1016/j.fm.2011.08.009
Andorrà I, Berradre M, Mas A, Esteve-Zarzoso B, Guillamón JM (2012) Effect of mixed culture fermentations on yeast populations and aroma profile. Food Sci Technol 49:8–13. https://doi.org/10.1016/j.lwt.2012.04.008
Anfang N, Brajkovich M, Goddard MR (2009) Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon Blanc. Aust J Grape Wine Res 15:1–8. https://doi.org/10.1111/j.1755-0238.2008.00031.x
Antalick G, Perello M-C, de Revel G (2010) Development, validation and application of a specific method for the quantitative determination of wine esters by headspace-solid-phase microextraction-gas chromatography–mass spectrometry. Food Chem 121:1236–1245. https://doi.org/10.1016/j.foodchem.2010.01.011
Arneborg N, Siegumfeldt H, Andersen GH, Nissen P, Daria VR, Rodrigo PJ, Glückstad J (2005) Interactive optical trapping shows that confinement is a determinant of growth in a mixed yeast culture. FEMS Microbiol Lett 245:155–159. https://doi.org/10.1016/j.femsle.2005.03.008
Bataillon M, Rico A, Sablayrolles J-M, Salmon J-M, Barre P (1996) Early thiamin assimilation by yeasts under enological conditions: impact on alcoholic fermentation kinetics. J Ferment Bioeng 82:145–150. https://doi.org/10.1016/0922-338X(96)85037-9
Beckner Whitener ME, Carlin S, Jacobson D, Weighill D, Divol B, Conterno L, Du Toit M, Vrhovsek U (2015) Early fermentation volatile metabolite profile of non-Saccharomyces yeasts in red and white grape must: a targeted approach. Food Sci Technol 64:412–422. https://doi.org/10.1016/j.lwt.2015.05.018
Beltran G, Torija MJ, Novo M, Ferrer N, Poblet M, Guillamón JM, Rozès N, Mas A (2002) Analysis of yeast populations during alcoholic fermentation: a six year follow-up study. Syst Appl Microbiol 25:287–293. https://doi.org/10.1078/0723-2020-00097
Bely M, Sablayrolles JM, Barre P (1990a) Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J Ferment Bioeng 70:246–252
Bely M, Sablayrolles JM, Barre P (1990b) Description of alcoholic fermentation kinetics—its variability and significance. Am J Enol Vitic 41:319–324
Branco P, Viana T, Albergaria H, Arneborg N (2015) Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int J Food Microbiol 205:112–118. https://doi.org/10.1016/j.ijfoodmicro.2015.04.015
Camarasa C, Sanchez I, Brial P, Bigey F, Dequin S (2011) Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147. https://doi.org/10.1371/journal.pone.0025147
Chasseriaud L, Miot-Sertier C, Coulon J, Iturmendi N, Moine V, Albertin W, Bely M (2015) A new method for monitoring the extracellular proteolytic activity of wine yeasts during alcoholic fermentation of grape must. J Microbiol Methods 119:176–179. https://doi.org/10.1016/j.mimet.2015.10.025
Cheraiti N, Guezenec S, Salmon JM (2005) Redox interactions between Saccharomyces cerevisiae and Saccharomyces uvarum in mixed culture under enological conditions. Appl Environ Microbiol 71:255–260. https://doi.org/10.1128/AEM.71.1.255-260.2005
Cheraiti N, Guezenec S, Salmon JM (2010) Very early acetaldehyde production by industrial Saccharomyces cerevisiae strains: a new intrinsic character. Appl Microbiol Biotechnol 86:693–700. https://doi.org/10.1007/s00253-009-2337-5
Chessel D, Dufour AB, Thioulouse J (2004) The ade4 package-I: one-table methods. R News 4:5–10
Ciani M, Mannazzu I, Marinangeli P, Clementi F, Martini A (2004) Contribution of winery-resident Saccharomyces cerevisiae strains to spontaneous grape must fermentation. Antonie Van Leeuwenhoek 85:159–164. https://doi.org/10.1023/B:ANTO.0000020284.05802.d7
Ciani M, Beco L, Comitini F (2006) Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int J Food Microbiol 108:239–245. https://doi.org/10.1016/j.ijfoodmicro.2005.11.012
Ciani M, Comitini F, Mannazzu I, Domizio P (2010) Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res 10:123–133. https://doi.org/10.1111/j.1567-1364.2009.00579.x
Clemente-Jimenez J, Mingorance-Cazorla L, Martínez-rodríguez S, Heras-Vázquez FJL, Rodríguez-Vico F (2004) Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol 21:149–155. https://doi.org/10.1016/S0740-0020(03)00063-7
Combina M, Elía A, Mercado L, Catania C, Ganga A, Martinez C (2005) Dynamics of indigenous yeast populations during spontaneous fermentation of wines from Mendoza, Argentina. Int J Food Microbiol 99:237–243. https://doi.org/10.1016/j.ijfoodmicro.2004.08.017
David V, Terrat S, Herzine K, Claisse O, Rousseaux S, Tourdot-Maréchal R, Masneuf-Pomarede I, Ranjard L, Alexandre H (2014) High-throughput sequencing of amplicons to monitor yeast biodiversity in must and wine. J Ind Microbiol Biotechnol 41:811–821
Divol B, Toit M, Duckitt E (2012) Surviving in the presence of sulphur dioxide: strategies developed by wine yeasts. Appl Microbiol Biotechnol 95:601–613. https://doi.org/10.1007/s00253-012-4186-x
Dufour M, Zimmer A, Thibon C, Marullo P (2013) Enhancement of volatile thiol release of Saccharomyces cerevisiae strains using molecular breeding. Appl Microbiol Biotechnol 97:5893–5905
Egli CM, Edinger WD, Mitrakul CM, Henick-Kling T (1998) Dynamics of indigenous and inoculated yeast populations and their effect on the sensory character of Riesling and chardonnay wines. J Appl Microbiol 85:779–789. https://doi.org/10.1046/j.1365-2672.1998.00521.x
Fleet GH (2003) Yeast interactions and wine flavour. Int J Food Microbiol 86:11–22
Garcia A, Carcel C, Dulau L, Samson A, Aguera E, Agosin E, Günata Z (2002) Influence of a mixed culture with Debaryomyces vanriji and Saccharomyces cerevisiae on the volatiles of a Muscat wine. J Food Sci 67:1138–1143
Garofalo C, Tristezza M, Grieco F, Spano G, Capozzi V (2016) From grape berries to wine: population dynamics of cultivable yeasts associated to "Nero di Troia" autochthonous grape cultivar. World J Microbiol Biotechnol 32:59. https://doi.org/10.1007/s11274-016-2017-4
Goddard MR (2008) Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology 89:2077–2082. https://doi.org/10.1890/07-2060.1
Grangeteau C, Gerhards D, Rousseaux S, von Wallbrunn C, Alexandre H, Guilloux-Benatier M (2015) Diversity of yeast strains of the genus Hanseniaspora in the winery environment: what is their involvement in grape must fermentation? Food Microbiol 50:70–77. https://doi.org/10.1016/j.fm.2015.03.009
Henick-Kling T, Edinger W, Daniel P, Monk P (1998) Selective effects of sulfur dioxide and yeast starter culture addition on indigenous yeast populations and sensory characteristics of wine. J Appl Microbiol 84:865–876
Hu K, Qin Y, Tao YS, Zhu XL, Peng CT, Ullah N (2016) Potential of glycosidase from non-Saccharomyces isolates for enhancement of wine aroma. J Food Sci 81:M935–M943. https://doi.org/10.1111/1750-3841.13253
Huxley C, Green ED, Dunham I (1990) Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6:236
Ingram M (1948) The germicidal effects of free and combined sulphur dioxide. J Chem Technol Biotechnol 67:18–21. https://doi.org/10.1002/jctb.5000670107
Jolly NP, Augustyn OPH, Pretorius IS (2003) The occurrence of non-Saccharomyces cerevisiae yeast species over three vintages in four vineyards and grape musts from four production regions of the western cape. S Afr J Enol Vitic 24:35–42
Jolly NP, Varela C, Pretorius IS (2014) Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 14:215–237. https://doi.org/10.1111/1567-1364.12111
Li S-S, Cheng C, Li Z, Chen J-Y, Yan B, Han B-Z, Reeves M (2010) Yeast species associated with wine grapes in China. Int J Food Microbiol 138:85–90. https://doi.org/10.1016/j.ijfoodmicro.2010.01.009
Marullo P, Bely M, Masneuf-Pomarede I, Pons M, Aigle M, Dubourdieu D (2006) Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res 6:268–279. https://doi.org/10.1111/j.1567-1364.2006.00034.x
Marullo P, Aigle M, Bely M, Masneuf-Pomarede I, Durrens P, Dubourdieu D, Yvert G (2007) Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res 7:941–952. https://doi.org/10.1111/j.1567-1364.2007.00252.x
Marullo P, Mansour C, Dufour M, Albertin W, Sicard D, Bely M, Dubourdieu D (2009) Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res 9:1148–1160
Masneuf-Pomarede I, Juquin E, Miot-Sertier C, Renault P, Laizet Y, Salin F, Alexandre H, Capozzi V, Cocolin L, Colonna-Ceccaldi B, Englezos V, Girard P, Gonzalez B, Lucas P, Mas A, Nisiotou A, Sipiczki M, Spano G, Tassou C, Bely M, Albertin W (2015) The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res 15:fov045. https://doi.org/10.1093/femsyr/fov045
Mateo JJ, Jimenez M, Huerta T, Pastor A (1991) Contribution of different yeasts isolated from musts of monastrell grapes to the aroma of wine. Int J Food Microbiol 14:153–160. https://doi.org/10.1016/0168-1605(91)90102-U
Meillon S, Viala D, Medel M, Urbano C, Guillot G, Schlich P (2010) Impact of partial alcohol reduction in Syrah wine on perceived complexity and temporality of sensations and link with preference. Food Qual Prefer 21:732–740. https://doi.org/10.1016/j.foodqual.2010.06.005
Nardi T, Corich V, Giacomini A, Blondin B (2010) A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiology 156:1686–1696. https://doi.org/10.1099/mic.0.036723-0
Nisiotou AA, Nychas GJ (2007) Yeast populations residing on healthy or botrytis-infected grapes from a vineyard in Attica, Greece. Appl Environ Microbiol 73:2765–2768. https://doi.org/10.1128/AEM.01864-06
Nissen P, Arneborg N (2003) Characterization of early deaths of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch Microbiol 180:257–263. https://doi.org/10.1007/s00203-003-0585-9
Padilla B, Gil JV, Manzanares P (2016) Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front Microbiol 7:411. https://doi.org/10.3389/fmicb.2016.00411
Pate JB, Lodge JP, Wartburg AF (1962) Effect of pararosaniline in the trace determination of sulfur dioxide. Anal Chem 34:1660–1662. https://doi.org/10.1021/ac60192a001
Perez-Ortin JE, Querol A, Puig S, Barrio E (2002) Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12:1533–1539. https://doi.org/10.1101/gr.436602
Pérez G, Fariña L, Barquet M, Boido E, Gaggero C, Dellacassa E, Carrau F (2011) A quick screening method to identify β-glucosidase activity in native wine yeast strains: application of Esculin Glycerol Agar (EGA) medium. World J Microbiol Biotechnol 27:47–55. https://doi.org/10.1007/s11274-010-0425-4
Pfliegler WP, Horvath E, Kallai Z, Sipiczki M (2014) Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiol Res 169:402–410. https://doi.org/10.1016/j.micres.2013.09.006
Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729. https://doi.org/10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B
Development Core Team R (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Renault PE, Albertin W, Bely M (2013) An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl Microbiol Biotechnol 97:4105–4119. https://doi.org/10.1007/s00253-012-4660-5
Renault P, Coulon J, de Revel G, Barbe JC, Bely M (2015) Increase of fruity aroma during mixed T. delbrueckii/S. cerevisiae wine fermentation is linked to specific esters enhancement. Int J Food Microbiol 207:40–48. https://doi.org/10.1016/j.ijfoodmicro.2015.04.037
Rodriguez ME, Lopes CA, Barbagelata RJ, Barda NB, Caballero AC (2010) Influence of Candida pulcherrima Patagonian strain on alcoholic fermentation behaviour and wine aroma. Int J Food Microbiol 138:19–25. https://doi.org/10.1016/j.ijfoodmicro.2009.12.025
Rojas V, Gil JV, Pinaga F, Manzanares P (2001) Studies on acetate ester production by non-Saccharomyces wine yeasts. Int J Food Microbiol 70:283–289
Romano P, Suzzi G, Comi G, Zironi R, Maifreni M (1997) Glycerol and other fermentation products of apiculate wine yeasts. J Appl Microbiol 82:615–618
Rossouw D, Bauer FF (2016) Exploring the phenotypic space of non-Saccharomyces wine yeast biodiversity. Food Microbiol 55:32–46. https://doi.org/10.1016/j.fm.2015.11.017
Sadoudi M, Tourdot-Marechal R, Rousseaux S, Steyer D, Gallardo-Chacon JJ, Ballester J, Vichi S, Guerin-Schneider R, Caixach J, Alexandre H (2012) Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol 32:243–253. https://doi.org/10.1016/j.fm.2012.06.006
Salvadó Z, Arroyo-López FN, Barrio E, Querol A, Guillamón JM (2011) Quantifying the individual effects of ethanol and temperature on the fitness advantage of Saccharomyces cerevisiae. Food Microbiol 28:1155–1161. https://doi.org/10.1016/j.fm.2011.03.008
Stitt M, Lilley RM, Gerhardt R, Heldt HW (1989) [32] Metabolite levels in specific cells and subcellular compartments of plant leaves. Meth Enzymol 174:518–552. https://doi.org/10.1016/0076-6879(89)74035-0
Swiegers JH, Pretorius IS (2005) Yeast modulation of wine flavor. Adv Appl Microbiol 57:131–175. https://doi.org/10.1016/S0065-2164(05)57005-9
Tofalo R, Schirone M, Torriani S, Rantsiou K, Cocolin L, Perpetuini G, Suzzi G (2012) Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol 29:18–26. https://doi.org/10.1016/j.fm.2011.08.014
Tominaga T, Furrer A, Henry R, Dubourdieu D (1998) Identification of new volatile thiols in the aroma of Vitis vinifera L. var. Sauvignon blanc wines. Flavour Fragr J 13:159–162. https://doi.org/10.1002/(sici)1099-1026(199805/06)13:3<159::aid-ffj709>3.0.co;2-7
Tristezza M, Vetrano C, Bleve G, Spano G, Capozzi V, Logrieco A, Mita G, Grieco F (2013) Biodiversity and safety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia Region, Italy. Food Microbiol 36:335–342. https://doi.org/10.1016/j.fm.2013.07.001
Varela C, Siebert T, Cozzolino D, Rose L, MCLean H, Henschke PA (2009) Discovering a chemical basis for differentiating wines made by fermentation with ‘wild’ indigenous and inoculated yeasts: role of yeast volatile compounds. Aust J Grape Wine Res 15:238–248
Viana F, Gil JV, Genovés S, Vallés S, Manzanares P (2008) Rational selection of non-Saccharomyces wine yeasts for mixed starters based on ester formation and enological traits. Food Microbiol 25:778–785. https://doi.org/10.1016/j.fm.2008.04.015
Wang C, Esteve-Zarzoso B, Mas A (2014) Monitoring of Saccharomyces cerevisiae, Hanseniaspora uvarum, and Starmerella bacillaris (synonym Candida zemplinina) populations during alcoholic fermentation by fluorescence in situ hybridization. Int J Food Microbiol 191:1–9. https://doi.org/10.1016/j.ijfoodmicro.2014.08.014
Wang C, Mas A, Esteve-Zarzoso B (2015) Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int J Food Microbiol 206:67–74. https://doi.org/10.1016/j.ijfoodmicro.2015.04.022
Zimmer A, Durand C, Loira N, Durrens P, Sherman DJ, Marullo P (2014) QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One 9:e86298. https://doi.org/10.1371/journal.pone.0086298
Zott K, Miot-Sertier C, Claisse O, Lonvaud-Funel A, Masneuf-Pomarede I (2008) Dynamics and diversity of non-Saccharomyces yeasts during the early stages in winemaking. Int J Food Microbiol 125:197–203. https://doi.org/10.1016/j.ijfoodmicro.2008.04.001
Zott K, Claisse O, Lucas P, Coulon J, Lonvaud-Funel A, Masneuf-Pomarede I (2010) Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol 27:559–567. https://doi.org/10.1016/j.fm.2010.01.006
Zott K, Thibon C, Bely M, Lonvaud-Funel A, Dubourdieu D, Masneuf-Pomarede I (2011) The grape must non-Saccharomyces microbial community: impact on volatile thiol release. Int J Food Microbiol 151:210–215. https://doi.org/10.1016/j.ijfoodmicro.2011.08.026
Funding
This work was founded by Nouvelle Aquitaine Region, Biolaffort, and Pernod-Ricard companies.
Author information
Authors and Affiliations
Contributions
WA, JC, VM, BCC, MBer, PM, and IMP conceived or designed the study. WA, AZ, CMS, MBel, and PM performed research, WA, JC, VM, BCC, MBel, PM, and IMP analyzed the data. WA, PM, and IMP wrote the paper.
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
Warren Albertin, Cécile Miot-Sertier, Marina Bely, and Isabelle Masneuf-Pomarede declare that they have no conflict of interest.
Adrien Zimmer, Margaux Bernard, Joana Coulon, Virginie Moine, and Philippe Marullo are affiliated with Biolaffort company, and Benoit Colonna-Ceccaldi is affiliated with Pernod-Ricard company.
Electronic supplementary material
Online Resource 1
(PDF 648 kb)
Rights and permissions
About this article
Cite this article
Albertin, W., Zimmer, A., Miot-Sertier, C. et al. Combined effect of the Saccharomyces cerevisiae lag phase and the non-Saccharomyces consortium to enhance wine fruitiness and complexity. Appl Microbiol Biotechnol 101, 7603–7620 (2017). https://doi.org/10.1007/s00253-017-8492-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8492-1