Abstract
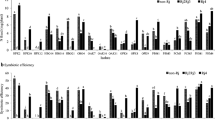
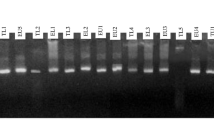
Analysis of genetic diversity among indigenous rhizobia and its symbiotic effectiveness with soybean cultivar is important for development of knowledge about rhizobial ecology. In India, little is known about the genetic resources and diversity of rhizobia nodulating soybean. Indigenous bradyrhizobia isolated from root nodules of soybean plants, collected from traditional cultivating regions of two states (Madhya Pradesh and Uttar Pradesh) of India, were screened for bacteriophage sensitivity to identify successful broad host range symbiotic effectivity. Of 172 rhizobial isolates, 91 showed sensitivities to eight lytic phages and form ten groups on the basis of sensitivity patterns. The genetic diversity of 23 isolates belonging to different phage groups was assessed along with that of strains USDA123 and USDA94 by the restriction fragment length polymorphism (RFLP) analysis of 16S rDNA, intergenic spacer (IGS) (16S–23S rDNA), and DnaK regions. RFLP analysis of 16S rDNA formed 5 groups, whereas 19 and 9 groups were revealed by IGS and the DnaK genes, respectively. The IGS regions showed many amplified polymorphic bands. Nine isolates which revealed high RFLP polymorphism in the abovementioned regions (16S rRNA, IGS, DnaK) were used for 16S rRNA sequence analyses. The results indicate that taxonomically, all isolates were related to Rhizobium etli, Bradyrhizobium spp., and Bradyrhizobium yuanmingense. The doubling time of isolates varied from 9 h (MPSR155) to 16.2 h (MPSR068) in YM broth. Five isolates which did not show cross infectivity with isolated phage strains were studied for symbiotic efficiency. All isolates showed broad host range symbiotic effectiveness forming effective nodules on Vigna mungo, Vigna radiata, Vigna unguiculata, and Cajanus cajan. The present study provides information on genetic diversity and host range symbiosis of indigenous soybean rhizobia typed by different phages.





Similar content being viewed by others
References
Abaidoo RC, Keyser HH, Singleton PW, Dashiell KE, Sanginga N (2007) Population size, distribution, and symbiotic characteristics of indigenous Bradyrhizobium spp. that nodulate TGx soybean genotypes in Africa. Appl Soil Ecol 35:57–67
Adams MH (1959) Bacteriophages. Wiley, New York
Ali FS, Loynachan TE, Hammad AMM, Aharchi Y (1998) Polyvirulent rhizobiophages from a soybean rhizosphere soil. Soil Biol Biochem 30:2171–2175
Amarger N (2001) Rhizobia in the field. Adv Agron 73:109–168
Appunu C, Dhar B (2006) Phage typing of indigenous soybean rhizobia and relationship of a phage group strains for their asymbiotic and symbiotic nitrogen fixation. Indian J Exp Biol 44:1006–1011
Appunu C, N’Zoue A, Laguerre G (2008) Genetic diversity of native bradyrhizobia isolated from soybeans (Glycin max L.) in different agricultural–ecological–climatic regions of India. Appl Environ Microbiol 74:5991–5996
Appunu C, N’Zoue A, Moulin L, Depret G, Laguerre G (2009) Vingna mungo, V. radiata and V. unguiculata plants sampled in different agronomical–ecological–climatic regions of India are nodulated by Bradyrhizobium yuanmingense. Syst Appl Microbiol 32:460–470
Barry T, Colleran G, Glennon M, Dunican LK, Gannon F (1991) The 16 s/23s ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl 1:51–56
Bromfield ESP, Tambong JT, Coutier S, Laguerre G, van Berkum P, Tran Thi TV, Assabgui R, Barran LR (2010) Ensifer, Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 156:505–520
Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H (2004) Phage–host interaction: an ecological perspective. J Bacteriol 186:3677–3686
Deosthali V, Akmanchi A, Salunke C (2005) Soybean agriculture in India, a spatial analysis. Trans Inst Indian Geogr 27:13–30
Doignon-Bourcier F, Willems A, Coopman R, Laguerre G, Gillis M, de Lajudie P (2000) Genotypic characterization of Bradyrhizobium strains nodulating small Senegalese legumes by 16S–23S rRNA intergenic gene spacers and amplified fragment length polymorphism fingerprint analyses. Appl Environ Microbiol 66:3987–3997
Estrell MJ, Munoz S, Soto MJ, Ruiz O, Sanjuan J (2009) Genetic diversity and host range of rhizobia nodulating Lotus tenuis in typical soils of the Salado River Basin (Argentina). Appl Environ Microbiol 75:1088–1098
Hashem FM, Angle JS, Ristiano PA (1986) Isolation and characterization of rhizobiophage specific for Rhizobium japonicum USDA 117. Can J Microbiol 32:326–329
Jaiswal SK, Dhar B (2010) Morphology and general characteristics of phages specific for Lens culinaris rhizobia. Biol Fertil Soils 46:681–687
Jaiswal SK, Dhar B (2011) Identification and assessment of symbiotic effectiveness of phage-typed Rhizobium leguminosarum strains on lentil (Lens culinaris Medik) cultivars. Curr Microbiol 62:1503–1509
Jensen MA, Straus N (1993) Effects of PCR conditions on the formation of heteroduplex and single-stranded DNA products in the amplification of bacterial ribosomal DNA spacer regions. PCR Methods Appl 3:186–194
Jensen MA, Webster JA, Straus N (1993) Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol 59:945–952
Jitacksorn S, Sadowsky MJ (2008) Nodulation gene regulation and quorum sensing control density-dependent suppression and restriction of nodulation in the Bradyrhizobium japonicum–soybean symbiosis. Appl Environ Microbiol 74:3749–3756
Kassen R, Rainey PB (2004) The ecology and genetics of microbial diversity. Annu Rev Microbiol 58:207–231
Kowalski M, Malek W, Czopska-Dolecka J, Szlachetka M (2004) The effect of rhizobiophages on Sinorhizobium meliloti–Medicago sativa symbiosis. Biol Fertil Soils 39:292–294
Lesley SM (1982) A bacteriophages typing system for Rhizobium meliloti. Can J Microbiol 28:180–189
Laguerre G, Mavingui P, Allard MR, Charnay MP, Louvrier P, Mazurier SI, Rigottier-Gois L, Amarger N (1996) Typing of rhizobia by PCR DNA fingerprinting and PCR- restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol 62:2029–2036
Lindstrom K, Lipsanen P, Kaijalainen S (1990) Stability of markers used for identification of two Rhizobium galgae inoculants strain after five years in the field. Appl Environ Microbiol 56:444–450
Lindstrom EK, Jarvis BDW, Lindstrom PE, Patel JJ (1982) DNA homology, phage-typing and cross nodulation studies of rhizobia infecting Galegae species. Can J Microbiol 29:781–789
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with Folin-phenol reagent. J Biol Chem 193:265–275
Mylona P, Pawlowski K, Bisseling T (1995) Symbiotic nitrogen fixation. Plant Cell 7:869–885
Nkot NL, Krasova-Wade T, Etoa FX, Sylla SN, Nwaga D (2008) Genetic diversity of rhizobia nodulating Arachis hypogaea L. in diverse land use systems of humid forest zone in Cameroon. Appl Soil Ecol 40:411–416
Ormeno-Orrillo E, Vinuesa P, Zuniga-Davila D, Martinez-Romero E (2006) Molecular diversity of native bradyrhizobia isolated from Lima bean (Phaseolus lunatus L.) in Peru. Syst Appl Microbiol 29:253–262
Pagan JD, Child JJ, Scowcroft WR, Gibson AH (1975) Nitrogen fixation by Rhizobium cultured on a defined medium. Nature 256:406–407
Rohlf FJ (1992) NTSYS-PC: numerical taxonomy and multivariate analysis system. Exeter Software, New York
Santamaria M, Corzo J, Leon-Barrios M, Gutierrez-Navarro AM (1997) Characterization and differentiation of indigenous rhizobia isolated from Canarian shrub legumes of agricultural and ecological interest. Plant Soil 190:143–152
Sarr A, Neyra M, Abdeljalil M, Houeibib O, Ndoye I, Oihabi A, Lesueur D (2005) Rhizobial populations in soils from natural Acacia senegal and Acacia nilotica forests in Mauritania and the Senegal River Valley. Microbiol Ecol 50:152–162
Shamseldin A, El-Saadani M, Sadowsky MJ, An CS (2009) Rapid identification and discrimination among Egyptian genotypes of Rhizobium leguminosarum bv. viciae and Sinorhizobium meliloti nodulating faba bean (Vicia faba L.) by analysis of nodC, ARDRA, and rDNA sequence analysis. Soil Biol Biochem 41:45–53
Somasegaran P, Hoben HJ (1994) Handbook of rhizobia. Methods in legume–Rhizobium technology. Springer, New York
Staniewski R (1970) Typing of Rhizobium by phages. Can J Microbiol 16:1003–1009
Suttle CA (2007) Marine viruses—major player in the global ecosystem. Nat Rev Microbiol 5:801–812
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thurman NP, Bromfield ESP (1988) Effect of variation within and between Madicago and Melilotus species on the composition and dynamics of indigenous populations of Rhizobium meliloti. Soil Biol Biochem 20:31–38
Vincent JM (1970) A manual for the practical study of root nodule bacteria, IBP Handbook No. 15. Blackwell, Oxford
Vinuesa P, Rademaker Jan LW, de Bruijn FJ, Werner D (1998) Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S–23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl Environ Microbiol 64:2096–2104
Wasike VW, Lesueur D, Wachira FN, Mungai NW, Mumera LM, Sanginga N, Mburu HN, Mugadi D, Wango P, Vanlauwe B (2009) Genetic diversity of indigenous Bradyrhizobium nodulating promiscuous soybean [Glycine max (L) Merr.] varieties in Kenya: impact of phosphorus and lime fertilization in two contrasting sites. Plant Soil 322:151–163
Yang JK, Zhang WT, Yuang TY, Zhou JC (2006) Genotypic characteristics of the rrn operon and genome of indigenous soybean bradyrhizobia in cropping zones of China. Can J Microbiol 52:968–976
Yang JK, Zhou JC (2008) Diversity, phylogeny and host specificity of soybean and peanut bradyrhizobia. Biol Fertil Soils 44:843–851
Acknowledgment
This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India, New Delhi, (grant no. BT/PR7251/AGR/21/209/2006).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jaiswal, S.K., Anand, A., Dhar, B. et al. Genotypic Characterization of Phage-Typed Indigenous Soybean Bradyrhizobia and Their Host Range Symbiotic Effectiveness. Microb Ecol 63, 116–126 (2012). https://doi.org/10.1007/s00248-011-9950-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9950-4