Abstract
MicroRNAs (miRNAs) are small RNAs that bind to mRNA targets and regulate their translation. A functional study of miRNAs and exploration of their utility as disease markers require miRNA extraction from biological samples, which contain large amounts of interfering compounds for downstream RNA identification and quantification. The most common extraction methods employ silica columns or the TRIzol reagent but give out low recovery for small RNAs probably due to their short strand lengths. Herein, we fabricated the titanium dioxide nanofibers using electrospinning to facilitate miRNA extraction and developed the optimal buffer conditions to improve miRNA recovery from biological matrices of cell lysate and serum. We found that our TiO2 fibers could obtain a recovery of 18.0 ± 3.6% for miRNA fibers while carrying out the extraction in the more complex medium of cell lysate, much higher than the 0.02 ± 0.0001% recovery from the commercial kit. The much improved extraction of miRNAs from our fibers could be originated from the strong coordination between TiO2 and RNA’s phosphate backbone. In addition, the binding, washing, and elution buffers judiciously developed in the present study can achieve selective extraction of small RNA shorter than 500 nucleotides in length. Our results demonstrate that TiO2 nanofibers can work as a valuable tool for extraction of miRNAs from biological samples with high recovery.

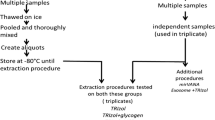
Schematic for extraction of small RNAs using TiO2 nanofibers






Similar content being viewed by others
References
Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40.
Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–3.
Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34.
Bekris LM, Leverenz JB. The biomarker and therapeutic potential of miRNA in Alzheimer’s disease. Neurodegener Dis Manag. 2015;5:61–74.
Alipoor SD, Adcock IM, Garssen J, Mortaz E, Varahram M, Mirsaeidi M, et al. The roles of miRNAs as potential biomarkers in lung diseases. Eur J Pharmacol. 2016;791:395–404.
Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46:953–60.
Cao Y, Griffith JF, Weisberg SB. The next-generation PCR-based quantification method for ambient waters: digital PCR. Methods Mol Biol. 2016;1452:113–30.
Al-Soud WA, Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–93.
Duy J, Koehler JW, Honko AN, Minogue TD. Optimized microRNA purification from TRIzol-treated plasma. BMC Genomics. 2015;16:95.
Rio DC, Ares M, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 2010;2010:pdb.prot5439.
Auffinger P, Bielecki L, Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–88.
Simões AES, Pereira DM, Amaral JD, Nunes AF, Gomes SE, Rodrigues PM, et al. Efficient recovery of proteins from multiple source samples after TRIzol(®) or TRIzol(®)LS RNA extraction and long-term storage. BMC Genomics. 2013;14:181.
Zhao X, Johnson JK. Simulation of adsorption of DNA on carbon nanotubes. J Am Chem Soc. 2007;129:10438–45.
Saha S, Sarkar P. Understanding the interaction of DNA-RNA nucleobases with different ZnO nanomaterials. Phys Chem Chem Phys. 2014;16:15355–66.
Park JS, Goo N-I, Kim D-E. Mechanism of DNA adsorption and desorption on graphene oxide. Langmuir. 2014;30:12587–95.
Hashemi E, Akhavan O, Shamsara M, Valimehr S, Rahighi R. DNA and RNA extractions from eukaryotic and prokaryotic cells by graphene nanoplatelets. RSC Adv. 2014;4:60720–8.
Wang F, Liu B, Huang P-JJ, Liu J. Rationally designed nucleobase and nucleotide coordinated nanoparticles for selective DNA adsorption and detection. Anal Chem. 2013;85:12144–51.
Madhugiri S, Sun B, Smirniotis PG, Ferraris JP, Balkus KJ. Electrospun mesoporous titanium dioxide fibers. Microporous Mesoporous Mater. 2004;69:77–83.
Lu C, Liu Y, Ying Y, Liu J. Comparison of mos2, WS2, and graphene oxide for DNA adsorption and sensing. Langmuir. 2017;33:630–7.
Kim J, Park S-J, Min D-H. Emerging approaches for graphene oxide biosensor. Anal Chem. 2017;89:232–48.
Pihlasalo S, Mariani L, Härmä H. Quantitative and discriminative analysis of nucleic acid samples using luminometric nonspecific nanoparticle methods. Nano. 2016;8:5902–11.
Jena PV, Safaee MM, Heller DA, Roxbury D. DNA-carbon nanotube complexation affinity and photoluminescence modulation are independent. ACS Appl Mater Interfaces. 2017;9:21397–405.
Vilela P, El-Sagheer A, Millar TM, Brown T, Muskens OL, Kanaras AG. Graphene oxide-upconversion nanoparticle based optical sensors for targeted detection of mRNA biomarkers present in Alzheimer’s disease and prostate cancer. ACS Sens. 2017;2:52–6.
Li F, Liu X, Zhao B, Yan J, Li Q, Aldalbahi A, et al. Graphene nanoprobes for real-time monitoring of isothermal nucleic acid amplification. ACS Appl Mater Interfaces. 2017;9:15245–53.
Bielicka-Daszkiewicz K, Voelkel A. Theoretical and experimental methods of determination of the breakthrough volume of SPE sorbents. Talanta. 2009;80:614–21.
Thingholm TE, Larsen MR. The use of titanium dioxide for selective enrichment of phosphorylated peptides. Methods Mol Biol. 2016;1355:135–46.
Eriksson AIK, Bartsch M, Bergquist J, Edwards K, Lind SB, Agmo Hernández V. On-target titanium dioxide-based enrichment for characterization of phosphorylations in the Adenovirus pIIIa protein. J Chromatogr A. 2013;1317:105–9.
Wakabayashi M, Kyono Y, Sugiyama N, Ishihama Y. Extended coverage of singly and multiply phosphorylated peptides from a single titanium dioxide microcolumn. Anal Chem. 2015;87:10213–21.
Piovesana S, Capriotti AL, Cavaliere C, Ferraris F, Iglesias D, Marchesan S, et al. New magnetic graphitized carbon black TiO2 composite for phosphopeptide selective enrichment in shotgun phosphoproteomics. Anal Chem. 2016;88:12043–50.
Li Q, Ning Z, Tang J, Nie S, Zeng R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J Proteome Res. 2009;8:5375–81.
Zhang X, Wang F, Liu B, Kelly EY, Servos MR, Liu J. Adsorption of DNA oligonucleotides by titanium dioxide nanoparticles. Langmuir. 2014;30:839–45.
Mondal K, Ali MA, Agrawal VV, Malhotra BD, Sharma A. Highly sensitive biofunctionalized mesoporous electrospun TiO(2) nanofiber based interface for biosensing. ACS Appl Mater Interfaces. 2014;6:2516–27.
Chen X, Mao SS. Titanium dioxide nanomaterials: synthesis, properties, modifications, and applications. Chem Rev. 2007;107:2891–959.
Vandeventer PE, Mejia J, Nadim A, Johal MS, Niemz A. DNA adsorption to and elution from silica surfaces: influence of amino acid buffers. J Phys Chem B. 2013;117:10742–9.
Li D, Xia Y. Fabrication of titania nanofibers by electrospinning. Nano Lett. 2003;3:555–60.
Pirzada T, Arvidson SA, Saquing CD, Shah SS, Khan SA. Hybrid silica-PVA nanofibers via sol-gel electrospinning. Langmuir. 2012;28:5834–44.
Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6:127.
Masotti A, Preckel T. Analysis of small RNAs with the Agilent 2100 Bioanalyzer. Nat Methods. 2006;3:iii–v.
Acknowledgments
We are thankful for the kind assistance from Mr. Joshua Belardes on the early stage of buffer optimization for DNA extraction with SiO2 fibers.
Funding
The authors acknowledge the support by the National Cancer Institute of the National Institutes of Health under Award Number R01CA188991 to WZ. This award also provided support (3R01CA188991-02S1) under the Research Supplements to Promote Diversity in Health-Related Research Program for LAJ. MAG was supported by MARCU-STAR training grant from NIH. SS and YM were supported by the Research in Science and Engineering (RISE) undergraduate summer research program at UC Riverside. JGC was supported by UC Riverside’s office of Undergraduate Education (UE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Informed consent
The serum samples were obtained from commercial sources with the individual information unknown to the researchers.
Additional information
Published in the topical collection celebrating ABCs 16th Anniversary.
Electronic supplementary material
ESM 1
(PDF 156 kb)
Rights and permissions
About this article
Cite this article
Jimenez, L.A., Gionet-Gonzales, M.A., Sedano, S. et al. Extraction of microRNAs from biological matrices with titanium dioxide nanofibers. Anal Bioanal Chem 410, 1053–1060 (2018). https://doi.org/10.1007/s00216-017-0649-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0649-3