Abstract
Cold-active bacteria are currently of great interest in biotechnology, and their genomic and physiological features have been extensively studied. One of the model psychrotolerant bacteria are Psychrobacter spp. Analysis of Arctic psychrophilic Psychrobacter sp. DAB_AL32B genome content provided an insight into its overall stress response, and genes conferring protection against various life-limiting factors (i.e., low temperature, increased ultraviolet radiation, oxidative stress and osmotic pressure) were recognized and described. Moreover, it was revealed that the strain carries a large plasmid pP32BP2. Its replication system was used for the construction of two novel shuttle vectors (pPS-NR—Psychrobacter-Escherichia coli-specific plasmid and pPS-BR—Psychrobacter-various Proteobacteria-specific plasmid) of an increased carrying capacity, which may be used for genetic engineering of Psychrobacter spp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Psychrobacter spp. belong to the Moraxellaceae family (Gammaproteobacteria). Bacteria of this genus are frequently isolated from various cold environments, including seawater, ice, permafrost and Arctic and Antarctic ornithogenic soils. Some strains (e.g., P. pulmonis and P. phenylpyruvicus) are considered to be opportunistic pathogens, as they are occasionally isolated from human patients, as well as from infected animals (Bowman et al. 1996; Bowman 2006; Ortiz-Alcantara et al. 2016).
Psychrobacter strains grow in temperatures between − 10 and 38 °C, and the majority of strains are psychrotolerants. Usually, Psychrobacter spp. are also recognized as halotolerants, since they are able to grow in the presence of 6.5% saline and above. Their ability to thrive in permanently cold and high-salt environments distinguish Psychrobacter spp. from their closest relatives, i.e., Acinetobacter and Moraxella (Maruyama et al. 2000; Garrity 2005; Bowman 2006).
According to the NCBI genome browser (http://ncbi.nlm.nih.gov/genome/browse), 77 genomes (including 65 drafts) and an additional 65 plasmid sequences of Psychrobacter spp. are available (22nd June 2018). Analysis of the Psychrobacter genomes revealed the presence of various adaptation mechanisms allowing their survival in extremely cold environments. Hence, psychrophilic Psychrobacter strains are model, cold-active bacteria useful for studying bacterial adaptation to extreme conditions (Bakermans et al. 2007; Zheng et al. 2007; Ayala-del-Rio et al. 2010; Moghadam et al. 2016; Dias et al. 2018). Moreover, they constitute a biotechnologically valuable source of cold-active enzymes, e.g., carbonic anhydrase and lipases (Zhang et al. 2007; Parra et al. 2008; Li et al. 2016; Lasa and Romalde 2017). It is also worth mentioning that, Psychrobacter spp. were reported to exhibit resistances to various heavy-metal ions, including arsenate, arsenite, copper, tellurite, and chromate (Dziewit et al. 2013; Arenas et al. 2014; Munoz-Villagran et al. 2018). Heavy-metal resistance may be beneficial in the light of a potential application of Psychrobacter strains in various bioremediation technologies.
Considering the application of Psychrobacter spp. or their enzymes in various biotechnologies, it was found that these bacteria are usually quite easy to cultivate under laboratory conditions, as they grow rapidly at 20–25 °C and undergo genetic recombination by conjugation and electroporation. There are reports concerning genetic modifications applying transposon mutagenesis and allele exchange carried out for Psychrobacter spp. (Bakermans et al. 2009; Jeong et al. 2013). Therefore, it is possible to use bacteria of this genus as biological factories, e.g., for synthesis of cold-adapted enzymes, which cannot be produced in commonly found mesophilic strains. However, there is an urgent need for novel vectors specific for Psychrobacter spp., which can be used for cloning of exogenous DNA and expression of proteins in that host.
It was previously reported that plasmids carrying ColE1- and p15a-type replication systems were stably maintained in P. arcticus 274-3 (Bakermans et al. 2009). Nonetheless, in our experiments with several cold-active Psychrobacter strains, we were unable to introduce (neither by electroporation nor triparental mating) a pABW1 vector containing a ColE1-type replication system. Therefore, the broad host range vector pBBR1 MCS-2 with pBBR1-type replication system was tested. Surprisingly, this replication system was also found to be inactive in tested Psychrobacter spp. These negative results encouraged us to construct novel, shuttle Psychrobacter–Escherichia coli-specific vectors.
So far, five such vectors have been made available. Four shuttle vectors (pPCV1-4), based on the pBGS18 (carrying E. coli-specific replication system originating from the pMB1 plasmid, i.e., high copy number replicon) or pWSK29 (carrying E. coli-specific replication system originating from the pSC101 plasmid, i.e., low copy-number replicon) plasmids, and carrying replication systems of the pP43BP3 and pP43BP4 plasmids originating from psychrophilic Psychrobacter sp. DAB_AL43B, were reported (Lasek et al. 2017). Additionally, a pUC-ORIT/rep vector, based on E. coli-specific pUC18 vector and pTAUp replication system originating from 1.9-kb plasmid of Antarctic Psychrobacter sp. TA144, was constructed (Tutino et al. 2000). It is noteworthy that all above-mentioned vectors were constructed applying replication systems of relatively small replicons (1.9–6.45 kb). Cloning of large fragments using vectors containing replication systems of small-size plasmids may result in an insert instability, as was shown for, e.g., vector pSUP106 with a replication system derived from 8.7-kb plasmid RSF1010 (Priefer et al. 1985). Therefore, we recognized the need for Psychrobacter-specific vectors based on replication systems of large plasmids, and therefore potentially having an increased carrying capacity.
In this study, the draft genome sequence of Psychrobacter sp. DAB_AL32B isolated from the Hornsund, Spitsbergen (Arctic) was obtained. The genome analyses provided brief insights into the stress adaptation mechanisms of the strain. Moreover, we found that the strain carries a large, approximately 60-kb, plasmid. Although this plasmid was not fully assembled during genome drafting, its replication system was used for the construction of two novel Psychrobacter-specific vectors that are suitable for cloning of large DNA fragments.
Materials and methods
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown on lysogeny broth (LB) medium (Sambrook and Russell 2001) at 22 °C (Psychrobacter spp.) or 37 °C (Escherichia coli DH5α). The medium was solidified by the addition of 1.5% (w/v) agar. Where necessary, the media were supplemented with X-gal, IPTG and antibiotics: kanamycin (20 µg/ml for Psychrobacter spp. or 50 µg/ml for E. coli) and rifampin (50 µg/ml).
DNA sequencing
The CTAB/lysozyme method was used for isolation of genomic DNA (Sambrook and Russell 2001). An Illumina TruSeq library was constructed following the manufacturer’s instructions. Sequencing was performed on an Illumina MiSeq instrument using the v3 chemistry kit. Sequence reads were filtered for quality and assembled using Newbler version 3.0 software with default settings (Roche, Basel, Switzerland), as it generated longest contigs (compared to other assembling tools). PCR products were cloned into the pABW1 and pBBR1 MCS-2 vectors and the resulting plasmids were used for DNA sequencing applying a dye terminator sequencing kit and an automated sequencer (ABI 377 PerkinElmer) (Applied Biosystems, Waltham, USA). Primer walking was employed to obtain the complete nucleotide sequence of the cloned PCR products.
All sequencing was performed in the DNA Sequencing and Oligonucleotide Synthesis Laboratory at the Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland).
DNA manipulations and introduction of plasmid DNA into bacterial cells
Plasmid DNA was isolated using a GeneMATRIX Plasmid Miniprep DNA Purification Kit (EURx, Gdansk, Poland) and alkaline lysis method (Birnboim and Doly 1979). Standard DNA manipulations were performed according to Sambrook and Russell (2001). PCR was performed in a Mastercycler (Eppendorf, Hamburg, Germany) using KAPA HiFi PCR Kit (KAPA polymerase with supplied components) (KAPA Biosystems, Cape Town, South Africa) and primer pair (the restriction enzyme sites are underlined) 32B_REPF—5′-ATACCGGTGCGAACCACTGTGAGTATTG-3′ and 32B_REPR—5′-ATACCGGTTTAATTCTATCGCCCGCCTG-3′. To obtain the PCR product, the following conditions were applied: 35 cycles (denaturation at 98 °C for 20 s, annealing at 61.4 °C for 60 s, extension at 72 °C for 30 s per 1 kb) preceded by 3-min denaturation at 95 °C and followed by 2-min extension at 72 °C.
The shuttle vectors were introduced into E. coli DH5α by transformation (Kushner 1978) and into Psychrobacter spp. via triparental mating. The triparental mating was conducted as follows: recipient Rifr-Psychrobacter strains were cultivated on LB agar plates for 1 day at 22 °C, then donor and helper strains were spread on a plate with pre-grown recipient strain and cultivated for 2 days at 22 °C. After 2 days of incubation, the bacteria were washed off the plate and suitable dilutions were plated on selective medium containing rifampin and kanamycin to select transconjugants carrying introduced Kmr-vector. The plates were cultivated for the next 3 days at 22 °C.
Plasmid stability assay
The pPS-NR plasmid was introduced into the DAB_AL12R and DAB_AL43BR strains via triparental mating. The presence of pPS-NR within analyzed strains was confirmed via alkaline lysis. Segregational stability of the pPS-NR vector in the DAB_AL12R and DAB_AL43BR strains was tested by replica plating, following growth under non-selective conditions for 30 generations, as described previously (Dziewit et al. 2007; Romaniuk et al. 2017).
Bioinformatics
The draft genome was automatically annotated using the NCBI Prokaryotic Genome Annotation Pipeline. Similarity searches were performed using the BLAST programs (Altschul et al. 1997). Secondary structures of proteins were predicted using InterPro (Jones et al. 2014) and Motif Scan (Pagni et al. 2007). For plasmids’ assembly from the draft genome, the plasmidSPAdes v. 3.12.0 tool (Antipov et al. 2016) was used. All programs were used with default settings.
Genome completeness was assessed by the presence/absence of bacterial orthologs according to the OrthoDB database using BUSCO (Simao et al. 2015). Genome contamination was determined using Taxoblast v. 1.1 (Dittami and Corre 2017) followed by manual evaluation of results. All contigs representing analyzed genome were splitted into sequences of 1000 bp length. Each DNA fragment was individually searched for homologous sequence in GenBank nr database (e value cut off 0.01) excluding genome of the analyzed DAB_AL32B strain. The Psychrobacter (taxon ID 497) was specified as taxon separating from other bacterial taxa.
PYANI (Pritchard et al. 2016) with ANIb method was used to calculate ANI values between the DAB_AL32B strain and each of 76 genomes of Psychrobacter spp.
16S rRNA gene phylogenetic analysis
Phylogenetic analysis was performed based on the comparison of partial 16S rRNA gene sequences of the DAB_AL32B strain, DAB_AL43B strain and strains representing 48 Psychrobacter species described to date. Sequences were aligned using the RDP Aligner tool (structure-aware multiple sequence aligner) available at the Ribosomal Database Project (RDP) website (Nawrocki and Eddy 2007). Phylogenetic tree was built using Tree Builder (with default settings) available at the RDP website. The 16S rRNA gene sequences used for phylogenetic analysis were collected from GenBank (NCBI). The GenBank sequences used for this analysis are as follows: P. adeliensis DSM 15333T (HE654007.1), P. aestuarii SC35 (EU939718.1), P. alimentarius JG-100 (AY513645.1), P. allis E2 (JX122558.1), P. aquaticus CMS 56 (NR_042206.1), P. aquimaris SW-210 (AY722804.1), P. arcticus 273-4 (NR_075054.1), P. arenosus R7T (AJ609273.1), P. celer SW-238 (NR_043225.1), P. ciconiae 176/10 (KM486054.1), P. cryohalolentis K5 (NR_075055.1), P. faecalis DSM 14664 (NR_118025.1), P. fjordensis BSw21516B (GQ358940.1), P. fozii NF23 (NR_025531.1), P. frigidicola DSM 12411 (NR_042222.1), P. fulvigenes KC 40 (NR_041688.1), P. glacialis DD43 (AJ539102.1), P. glaciei BIc20019 (NR_148850.1), P. glacincola DSM 12194 (NR_042076.1), P. halophilus DD2 (AJ539103.1), P. immobilis ATCC 43116 (NR_118808.1), P. jeotgali YKJ-103 (NR_025205.1), P. luti NF11 (NR_025532.1), P. lutiphocae IMMIB L-1110 (NR_044602.1), P. marincola KMM 277 (NR_025458.1), P. maritimus Pi2-20 (NR_027225.1), P. meningitidis SBA4 (KR091838.1), P. muriicola 2pS (NR_114669.1), P. namhaensis SW-242 (AY722805.1), P. nivimaris 88/2–7 (AJ313425.1), P. oceani 4k5 (AB910522.1), P. okhotskensis MD17 (NR_024806.1), P. pacificensis NBRC 103191 (NR_114238.1), P. pasteuri CIP110853 (KY292376.1), P. phenylpyruvicus ATCC 23333 (NR_118815.1), P. piechaudii CIP110854 (KY292375.1), P. piscatorii T-3-2 (NR_112807.1), P. piscidermidis 45 (FJ613616.1), P. pocilloporae S6-60 (KT444699.2), P. proteolyticus HAMBI 2948 (LT899990.1), P. psychrophilus BBDP29 (DQ337513.1), P. pulmonis CCUG 46240 (NR_118026.1), P. salsus DD 48 (NR_042166.1), P. sanguinis 13983 (HM212668.1), P. submarinus KMM 225 (AJ309940.1), P. urativorans DSM 14009T (AJ609555.1), P. vallis CMS 39 (AJ584832.1), P. cibarius JG-219 (AY639871.1), Psychrobacter sp. DAB_AL32B (JF714884.1) and Psychrobacter sp. DAB_AL43B (JF714885.1).
Nucleotide sequence accession number
The Psychrobacter sp. DAB_AL32B draft genome sequence has been deposited in the GenBank (NCBI) database under accession numbers NEXU01000001-NEXU01000218 and the sequences of vectors constructed in this study under accession numbers MH539767 (pPS-BR) and MH539768 (pPS-NR).
Results and discussion
General features of the Psychrobacter sp. DAB_AL32B genome
The estimated size of the DAB_AL32B genome is 3,211,529 bp, which is similar to the size of other Psychrobacter spp. genomes available in the NCBI database (the average length of the Psychrobacter genome is 3.07 Mb). An assembly of the DAB_AL32B draft genome resulted in 37 scaffolds composed of 218 contigs. The average GC content is 41.9%, which is typical for Psychrobacter spp. (the average value is 43.26%). An automatic annotation of the DAB_AL32B genome performed applying the NCBI Prokaryotic Genome Annotation Pipeline resulted in 2,799 predicted genes with an average length of 973.43 bp, which covers about 80.08% of the genome. Additionally, 41 tRNA genes were identified (Table S1, Supplementary materials).
To estimate the completeness of the obtained draft genome, a BUSCO analysis, using Gammaproteobacteria BUSCO set (containing 452 BUSCO groups), was performed. The gene set predicted within the DAB_AL32B genome contains 94.3% of genes present in an applied BUSCO set (92.5% complete and 1.8% fragmented genes), which suggests that the genome assembly is complete or very close to completeness.
To examine if obtained draft genome is contaminated with other genomic sequences, an analysis using Taxoblast was performed (Table S2, Supplementary material). This analysis revealed that 30 out of 218 contigs were classified as possible contaminants or did not exhibit significant homology to sequences available in the GenBank database. These contigs were manually examined. Putative prophage was found within contig00038. Seven contigs (contig00009, contig00020, contig00096, contig00102, contig00104, contig00122, and contig00128) mostly exhibited homology to Psychrobacter genomes, with only short fragments homologous to genomes of phylogenetically closely related Moraxella and Acinetobacter. Other two contigs (contig00048 and contig00081) exhibit similarity to Psychrobacter genomes in majority of their fragments, while the remaining fragments are either homologous to Acinetobacter, Moraxella, and other unrelated bacteria, or not homologous to any sequence in the nr database. However, BLASTx search revealed significant similarity to proteins encoded within most of these fragments to Psychrobacter and Acinetobacter proteins. For three other contigs (contig00140, contig00162 and contig00205) the best BLAST hits were sequences from either Acinetobacter or Moraxella; however, they showed comparable similarity to Psychrobacter genomes. One contig (contig00082) was mistakenly automatically classified as a contaminant, because of a non-significant hit to zebrafish genome in low-complexity region. As a result of above analysis, these 15 contigs were recategorized as non-contaminant sequences. Seven other contigs (contig00002, contig00150, contig00177, contig00184, contig00193, contig00199, and contig00218) exhibited no significant homology to sequences available in the GenBank database (total length 2,557 bp, which comprises 0.0008% of the draft genome), therefore it is unclear if these are contaminants or not. Finally, only nine contigs (total length 4,558 bp, 0.0014% of the draft genome) (contig00134, contig00143, contig00146, contig00174, contig00181, contig00195, contig00196, contig00197, and contig00210) may be considered as potential contaminants, as they do not show similarities to neither Psychrobacter sequences nor closely related bacteria.
Phylogenetic analysis of Psychrobacter sp. DAB_AL32B
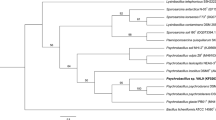
Previous phylogenetic analysis (Dziewit et al. 2013), based on the comparison of partial 16S rDNA sequences, showed that the DAB_AL32B strain is closely related to Psychrobacter frigidicola DSM 12411. As several new Psychrobacter species have been described since the last analysis, we performed another analysis with dataset extended on 14 new Psychrobacter-type strains (Fig. 1). Same as previously, the DAB_AL32B strain was clustered with P. frigidicola DSM 12411.
Phylogenetic tree for 16S rDNA sequences of Psychrobacter spp. The DAB_AL32B strain is denoted by the bold text. The statistical support for the internal nodes was determined by 1000 bootstrap replicates and values of > 50% are shown. The scale bar represents 0.004 substitutions per nucleotide position. The tree was not rooted
For more precise classification of DAB_AL32B at the species level, the ANI analysis, based on whole-genome comparison between various Psychrobacter strains, was performed. The DAB_AL32B genome was compared to each of 76 Psychrobacter genomes available (18th July 2018) in NCBI database (Table S3, Supplementary materials). Unfortunately, the genome of P. frigidicola DSM 12411 or some other representative of this species, is not publicly available. The phylogenomic analysis revealed that the DAB_AL32B strain is most related with Psychrobacter sp. DAB_AL43B (ANI value 95.672). However, to predict if these strains may be considered as representatives of novel Psychrobacter species, the calculation of the ANI value with P. frigidicola is needed.
Genes involved in the stress response of the DAB_AL32B strain
Various adaptive mechanisms enable survival of psychrophilic bacteria in Arctic. Identification and analysis of genes encoding various stress response proteins of psychrophiles is important for studying their biology and adaptation, as well as for proper recognition of their biotechnological potential (D’Amico et al. 2006; Casanueva et al. 2010; De Maayer et al. 2014; Santiago et al. 2016). In its natural environment, Psychrobacter sp. DAB_AL32B had to cope not only with low temperatures, but also increased ultraviolet (UV) radiation, osmotic pressure and oxidative stress. The genes involved in the response to oxidative, osmotic, and cold shock encoded within the DAB_AL32B genome are listed in Table 2.
Analysis of the DAB_AL32B genome showed that the most represented group of genes encoding stress response proteins is linked with oxidative stress, including protection against reactive oxygen species (ROS) formed in the cell. Increased ROS formation in cells may occur as a result of (i) depletion of the UV-protective ozone layer in the Arctic region (He and Hader 2002; Dugo et al. 2012) and (ii) permanent low temperatures, which increases the solubility of oxygen in water (Casanueva et al. 2010; Baez and Shiloach 2014). Cells exposed to elevated oxygen concentrations accumulate ROS, formed as byproducts of aerobic metabolism. Oxidants are highly reactive molecules and trigger damage in cellular components. They cause the oxidation of amino acids, which leads to protein fragmentation, the formation of aggregates and proteolysis (Cabiscol et al. 2000). DNA modifications caused by ROS include single- and double-strand breaks, bases modifications and cross-linking with proteins, leading to the mutation and rearrangement of DNA (Cabiscol et al. 2000; Jena 2012). Moreover, as a result of oxidation (induced by ROS and facilitated by Fe2+ ions), lipids cross-linking with proteins and disturbances to membrane structure affecting its fluidity are observed (Cabiscol et al. 2000; Repetto 2012). Bacteria may control ROS formation and protect themselves from oxidative stress applying various mechanisms, including scavenging ROS, maintaining a strong reducing environment in the cytosol and direct protection of vulnerable molecules (Cabiscol et al. 2000; Imlay 2013).
Within the DAB_AL32B genome, 11 genes encoding enzymes directly responsible for a diminishing amount of toxic ROS were found: catalases (3 genes), peroxidase (1), superoxide dismutase (1), peroxiredoxin (2), rubredoxin (1) and glutaredoxins (3) (Table 2). Additionally, 13 genes encoding enzymes involved in glutathione metabolism are present within the genome. Glutathione is a low-molecular weight thiol, protecting the cell from oxidative stress induced by peroxides (Masip et al. 2006).
Oxygen toxicity is increased by an excess of an intracellular iron in its reduced form (Fe2+). Hydroxyl radicals are then produced via Fenton reaction (Touati 2000). Therefore, it is essential to maintain a low and safe level of intracellular concentration of these ions. Proteins belonging to the ferritin superfamily are responsible for iron storage in its nontoxic oxidized form (Castruita et al. 2006; Arosio et al. 2009; Calhoun and Kwon 2011). Within the DAB_AL32B genome, two genes encoding Dps-like DNA-binding proteins were found (Table 2). These proteins contain both DNA-binding and ferroxidase domains and their proposed biological function is DNA protection from oxidation (via sequestration and oxidation of Fe2+ ions) and formation of very stable complexes with DNA (Castruita et al. 2006).
The annual temperature in the Spitsbergen ranges between − 35.9 and 13.5 °C (Nowosielski 2004; Przybylak and Arazny 2006). Therefore, bacteria of the genus Psychrobacter inhabiting this region have to cope with permanent cold. Low temperatures significantly influence cell functioning by altering protein conformation (and, in consequence, its activity), decreasing gene expression and membrane fluidity (Zecchinon et al. 2001; Chattopadhyay 2006; D’Amico et al. 2006). Psychrobacter sp. DAB_AL32B possesses genes encoding three proteins, i.e., CspA, CspC, and CspE, belonging to the family of cold-shock proteins (Table 2). Low temperatures induce stabilisation of secondary structures in RNA, which suppresses gene expression by blocking RNA polymerase and ribosomes (Phadtare and Severinov 2010). Csp proteins act as chaperones and resolve secondary structures in RNA, which prevents premature transcription termination and blocking of translation (Phadtare et al. 1999; Kaufman-Szymczyk et al. 2009; Phadtare and Severinov 2010; Song et al. 2012; Keto-Timonen et al. 2016).
To cope with the permanent cold, but also with the osmotic stress, bacteria intake compatible solutes (e.g., proline, choline and glycine betaine) from the environment or synthesise them in elevated quantities. Compatible solutes increase the stability of proteins and cell membranes without interfering with cellular function (Ziegler et al. 2010). The DAB_AL32B strain has five genes encoding osmolyte transporters belonging to the BCCT family (betaine/carnitine/choline transporter) and two genes encoding subunits of the ABC-type proline/glycine betaine transporter. In addition, four enzymes, involved in glycine betaine and choline synthesis, were found (Table 2).
Insight into plasmidome of Psychrobacter sp. DAB_AL32B
As reported previously, the DAB_AL32B strain harbours small (4.6-kb), cryptic plasmid pP32BP1 (Dziewit et al. 2013). Moreover, based on the alkaline lysis results, we found that the strain most probably carries also a large, approximately 60-kb, plasmid, named pP32BP2. It was also partially confirmed by the genome drafting. Using plasmidSPAdes, three contigs, i.e.: contig00102 (GenBank: NEXU01000102), contig00103 (GenBank: NEXU01000103), and contig00104 (GenBank: NEXU01000104) were predicted as potential fragments of the pP32BP2 plasmid. Obtaining the complete genomic sequence of this plasmid is the aim of our further studies. Within the contig00104, genes encoding proteins involved in plasmid replication and partitioning were found.
The replication initiation protein RepA (GenBank: OXL18943) of pP32BP2 shows 76% identity with RepB protein (GenBank: AFD62164) of pP62BP1 of Psychrobacter sp. DAB_AL62B (Lasek et al. 2012). Analysis of the RepA sequence revealed the existence of a leucine zipper motif (LVAKSNDLIVASYELTRNEQRL, residues 5–26, leucines are underlined) within the N-terminal part of this protein and winged helix DNA-binding domain (residues 1–136 and 140–233). Upstream to the repA gene, the putative origin of replication (oriV) was found (position 33,081−33,234 in the contig00104). It is composed of five 20-bp-long direct repeats DR1.1-1.5 (5′-ATAACACTAAATGATGTGGG-3′) followed by a pair of 19-bp long inverted repeats, IR1 and IR2 (5′-AACACAACGTATAACAATA-3′ and 5′-TATTGTTATACGTTGTGTT-3′, respectively) (Fig. 2).
Genetic organization of the maintenance module of the pP32BP2 plasmid. Grey arrows indicate genes and their transcriptional orientation, dots indicate location of oriV and parS sites, thin black arrows (above the DNA sequence) indicate location of direct and/or inverted repeats within the oriV and parS sites. The start codons of the parA and repA genes are in frames. DR direct repeat, IR inverted repeat, oriV origin of plasmid replication, parA gene encoding partitioning protein A, parB gene encoding partitioning protein B, parS partitioning centromer-like site, repA gene encoding replication protein
Moreover, within the predicted pP32BP2-contig, genes encoding a pair of partitioning proteins, ParA and ParB (GenBank: OXL18944 and OXL18945, respectively), and centromer-like sequence parS, were found. ParA contains the ParA domain (residues 4-182) and shows 93% identity to ParA-like protein (GenBank: ABE76262) encoded within the plasmid 1 of Psychrobacter cryohalolentis K5. Within the ParB protein, the ParG domain, specific to the ParB-like proteins of, e.g., Salmonella enterica plasmid TP228 and Pseudomonas alcaligenes plasmid pRA2, was found (Hayes 2000). The predicted ParB protein shows 90% identity to the hypothetical protein (GenBank: ALF60955) of the plasmid 5 of Psychrobacter urativorans R10.10B. The putative parS centomer-like sequence is located upstream to the parA gene. It consists of eight direct repeats DR2.1-2.8 (5′-(A/T)ATACTCA-3′) (Fig. 2). Similar parS site organization was reported for the pP62BP1 plasmid (Lasek et al. 2012, 2017).
Construction of novel Psychrobacter-specific shuttle vectors
Identification of the replication module of a relatively large (approx. 60 kb) plasmid pP32BP2, encouraged us to use this system for the construction of novel Psychrobacter-specific cloning vectors. Currently, all available Psychrobacter-specific vectors are based on replication systems of small (not exceeding 6.4 kb) plasmids, which may exclude their application as molecular tools for cloning of large genetic modules. Since Psychrobacter spp. recently became a model bacterium representing cold-active microorganisms (Bakermans et al. 2007, 2009; Bergholz et al. 2009; Bakermans 2018), we recognized an urgent need for development of novel Psychrobacter-specific vectors with an increased carrying capacity.
In this study, two novel molecular tools, suitable for genetic engineering in Psychrobacter spp., were constructed (Fig. 3). The PCR-amplified (1.8-kb) replication system of the pP32BP2 plasmid was cloned into the PfoI site of the pABW1 vector. This enabled construction of a narrow host range Psychrobacter-E. coli shuttle vector, pPS-NR (6,250 bp). Its functionality was confirmed by introducing it (via triparental mating) into two Psychrobacter spp. strains, DAB_AL12R and DAB_AL43BR. Vector stability was tested in both these strains, and after approximately 30 generations of growth without antibiotic selection pressure, plasmid pPS-NR was found in 21% and 61% of the host cells, respectively (Fig. 4).
Genetic organization of the shuttle vectors pPS-NR and pPS-BR. Grey arrows indicate genes and their transcriptional orientation: aphA kanamycin resistance gene, lacZ’ partial β-galactosidase gene, mob relaxase gene enabling mobilization to conjugal transfer, orf gene encoding conserved hypothetical protein, rep gene encoding replication initiation protein of the pBBR1 MCS-2 vector, repA gene encoding replication initiation protein of the pP32BP2 plasmid. The ColE1-type origin (ori) of replication of the pABW1 vector and MCSs of both plasmids are indicated
In the course of our preliminary analyses, we surprisingly revealed that the commonly-used broad host range pBBR1 MCS-2 vector carrying the pBBR1-type replication system functional in a variety of Gram-negative bacteria, including: Agrobacterium tumefaciens, E. coli, Bordetella pertusis, Klebsiella pneumoniae, Pseudomonas aeruginosa, Pseudomonas putida, Pseudomonas stutzeri, Rhizobium meliloti, and Vibrio cholerae (Szpirer et al. 2001) is not able to replicate in tested Psychrobacter spp. As we are planning to conduct functional analyses of selected Psychrobacter sp. DAB_AL32B genes in various members of Proteobacteria, we constructed the second shuttle vector based on the above-mentioned broad host range plasmid pBBR1 MCS-2.
For the construction of this vector, the PCR-amplified (1.8-kb) DNA fragment containing the replication system of the pP32BP2 plasmid was cloned within PfoI site of the pBBR1 MCS-2 vector. This resulted in construction of a unique Psychrobacter-various Proteobacteria shuttle vector, pPS-BR (6929 bp). The vector was successfully introduced via triparental mating into Psychrobacter sp. DAB_AL12R and DAB_AL43BR. To analyse the carrying capacity of the pPS-BR vector, we cloned and successfully introduced to Psychrobacter spp. two relatively large restriction fragments of the DAB_AL32B genome, of a length of 5.8 kb (approx. the size of the vector) and 12.7 kb (approx. two times bigger than the vector), respectively.
Both vectors, pPS-NR and pPS-BR, carry: (i–ii) two replication systems, which enable their replication in Psychrobacter spp. and E. coli (pPS-NR) or various Proteobacteria (pPS-BR), (iii) the system enabling mobilization to conjugal transfer, (iv) aphA gene-conferring resistance to kanamycin, (v) multiple cloning site (MCS) and (vi) a selection marker (lacZ’ gene) enabling blue-white screening of clones in E. coli (Fig. 3). The obtained vectors are novel and convenient tools for conducting genetic manipulations in Psychrobacter spp. and other Proteobacteria species.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA (2016) plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387
Arenas FA, Pugin B, Henríquez NA, Arenas-Salinas MA, Diaz-Vasquez WA, Pozo MF, Munoz CM, Chasteen TG, Perez-Donoso JM, Vasquez CC (2014) Isolation, identification and characterization of highly tellurite-resistant, tellurite-reducing bacteria from Antarctica. Polar Sci 8:40–52
Arosio P, Ingrassia R, Cavadini P (2009) Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta 1790:589–599
Ayala-del-Rio HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW, Di Bartolo G, Hauser L, Land M, Bakermans C, Rodrigues D, Klappenbach J, Zarka D, Larimer F, Richardson P, Murray A, Thomashow M, Tiedje JM (2010) The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol 76:2304–2312
Baez A, Shiloach J (2014) Effect of elevated oxygen concentration on bacteria, yeasts, and cells propagated for production of biological compounds. Microb Cell Fact 13:181
Bakermans C (2018) Adaptations to marine versus terrestrial low temperature environments as revealed by comparative genomic analyses of the genus Psychrobacter. FEMS Microbiol Ecol 94:fiy102
Bakermans C, Tollaksen SL, Giometti CS, Wilkerson C, Tiedje JM, Thomashow MF (2007) Proteomic analysis of Psychrobacter cryohalolentis K5 during growth at subzero temperatures. Extremophiles 11:343–354
Bakermans C, Sloup RE, Zarka DG, Tiedje JM, Thomashow MF (2009) Development and use of genetic system to identify genes required for efficient low-temperature growth of Psychrobacter arcticus 273-4. Extremophiles 13:21–30
Bartosik D, Bialkowska A, Baj J, Wlodarczyk M (1997) Construction of mobilizable cloning vectors derived from pBGS18 and their application for analysis of replicator region of a pTAV202 mini-derivative of Paracoccus versutus pTAV1 plasmid. Acta Microbiol Pol 46:387–392
Bergholz PW, Bakermans C, Tiedje JM (2009) Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol 191:2340–2352
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucl Acid Res 7:1513–1523
Bowman JP (2006) The genus Psychrobacter. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 920–930
Bowman JP, Cavanagh J, Austin JJ, Sanderson K (1996) Novel Psychrobacter species from Antarctic ornithogenic soils. Int J Syst Bacteriol 46:841–848
Cabiscol E, Tamarit J, Ros J (2000) Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8
Calhoun LN, Kwon YM (2011) Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol 110:375–386
Casanueva A, Tuffin M, Cary C, Cowan DA (2010) Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol 18:374–381
Castruita M, Saito M, Schottel PC, Elmegreen LA, Myneni S, Stiefel EI, Morel FM (2006) Overexpression and characterization of an iron storage and DNA-binding Dps protein from Trichodesmium erythraeum. Appl Environ Microbiol 72:2918–2924
Chattopadhyay MK (2006) Mechanism of bacterial adaptation to low temperature. J Biosci 31:157–165
D’Amico S, Collins T, Marx JC, Feller G, Gerday C (2006) Psychrophilic microorganisms: challenges for life. EMBO Rep 7:385–389
De Maayer P, Anderson D, Cary C, Cowan DA (2014) Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep 15:508–517
Dias LM, Folador ARC, Oliveira AM, Ramos RTJ, Silva A, Barauna RA (2018) Genomic architecture of the two cold-adapted genera Exiguobacterium and Psychrobacter: evidence of functional reduction in the Exiguobacterium antarcticum B7 genome. Genome Biol Evol 10:731–741
Ditta G, Stanfield S, Corbin D, Helinski DR (1980) Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA 77:7347–7351
Dittami SM, Corre E (2017) Detection of bacterial contaminants and hybrid sequences in the genome of the kelp Saccharina japonica using Taxoblast. PeerJ 5:e4073
Dugo MA, Han F, Tchounwou PB (2012) Persistent polar depletion of stratospheric ozone and emergent mechanisms of ultraviolet radiation-mediated health dysregulation. Rev Environ Health 27:103–116
Dziewit L, Jazurek M, Drewniak L, Baj J, Bartosik D (2007) The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J Bacteriol 189:1983–1997
Dziewit L, Cegielski A, Romaniuk K, Uhrynowski W, Szych A, Niesiobedzki P, Zmuda-Baranowska MJ, Zdanowski MK, Bartosik D (2013) Plasmid diversity in arctic strains of Psychrobacter spp. Extremophiles 17:433–444
Garrity G (2005) Bergey’s manual of systematic bacteriology: volume two: the proteobacteria, part C. Springer, New York
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Hayes F (2000) The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol Microbiol 37:528–541
He YY, Hader DP (2002) Involvement of reactive oxygen species in the UV-B damage to the cyanobacterium Anabaena sp. J Photochem Photobiol B 66:73–80
Imlay JA (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454
Jena NR (2012) DNA damage by reactive species: mechanisms, mutation and repair. J Biosci 37:503–517
Jeong H-J, Lee H, Hong SG, Cho J-C, Lee HK, Lee YK (2013) Transposon mutagenesis of Psychrobacter cryohalolentis PAMC 21807 by tri-parental conjugation. Adv Polar Sci 24:223–230
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240
Kaufman-Szymczyk A, Wojtasik A, Parniewski P, Bialkowska A, Tkaczuk K, Turkiewicz M (2009) Identification of the csp gene and molecular modelling of the CspA-like protein from Antarctic soil-dwelling psychrotrophic bacterium Psychrobacter sp. B6. Acta Biochim Pol 56:63–69
Keto-Timonen R, Hietala N, Palonen E, Hakakorpi A, Lindstrom M, Korkeala H (2016) Cold shock proteins: a minireview with special emphasis on Csp-family of enteropathogenic Yersinia. Front Microbiol 7:1151
Kovach ME, Phillips RW, Elzer PH, Roop RM II, Peterson KM (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802
Kushner SR (1978) An improved method for transformation of E. coli with ColE1 derived plasmids. In: Boyer HB, Nicosia S (eds) Genetic engineering, Elsevier/North-Holland, Amsterdam, pp 17–23
Lasa A, Romalde JL (2017) Genome sequence of three Psychrobacter sp. strains with potential applications in bioremediation. Genom Data 12:7–10
Lasek R, Dziewit L, Bartosik D (2012) Plasmid pP62BP1 isolated from an Arctic Psychrobacter sp. strain carries two highly homologous type II restriction-modification systems and a putative organic sulfate metabolism operon. Extremophiles 16:363–376
Lasek R, Dziewit L, Ciok A, Decewicz P, Romaniuk K, Jedrys Z, Wibberg D, Schluter A, Puhler A, Bartosik D (2017) Genome content, metabolic pathways and biotechnological potential of the psychrophilic Arctic bacterium Psychrobacter sp. DAB_AL43B, a source and a host of novel Psychrobacter-specific vectors. J Biotechnol 263:64–74
Li M, Zhu X, Wilkinson S, Huang M, Achal V (2016) Complete genome sequence of carbonic anhydrase producing Psychrobacter sp. SHUES1. Front Microbiol 7:1442
Maruyama A, Honda D, Yamamoto H, Kitamura K, Higashihara T (2000) Phylogenetic analysis of psychrophilic bacteria isolated from the Japan Trench, including a description of the deep-sea species Psychrobacter pacificensis sp. nov. Int J Syst Evol Microbiol 50:835–846
Masip L, Veeravalli K, Georgiou G (2006) The many faces of glutathione in bacteria. Antioxid Redox Signal 8:753–762
Moghadam MS, Albersmeier A, Winkler A, Cimmino L, Rise K, Hohmann-Marriott MF, Kalinowski J, Ruckert C, Wentzel A, Lale R (2016) Isolation and genome sequencing of four Arctic marine Psychrobacter strains exhibiting multicopper oxidase activity. BMC Genom 17:117
Munoz-Villagran CM, Mendez KN, Cornejo F, Figueroa M, Undabarrena A, Morales EH, Arenas-Salinas M, Arenas FA, Castro-Nallar E, Vasquez CC (2018) Comparative genomic analysis of a new tellurite-resistant Psychrobacter. strain isolated from the Antarctic Peninsula. PeerJ 6:e4402
Nawrocki EP, Eddy SR (2007) Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput Biol 3:e56
Nowosielski L (2004) Klimat Spitsbergenu. Gazeta Obserwatora IMGW 2:14–17
Ortiz-Alcantara JM, Segura-Candelas JM, Garces-Ayala F, Gonzalez-Duran E, Rodriguez-Castillo A, Alcantara-Perez P, Wong-Arambula C, Gonzalez-Villa M, Leon-Avila G, Garcia-Chequer AJ, Diaz-Quinonez JA, Mendez-Tenorio A, Ramirez-Gonzalez JE (2016) Fatal Psychrobacter sp. infection in a pediatric patient with meningitis identified by metagenomic next-generation sequencing in cerebrospinal fluid. Arch Microbiol 198:129–135
Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L (2007) MyHits: improvements to an interactive resource for analyzing protein sequences. Nucl Acids Res 35:W433–W437
Parra LP, Reyes F, Acevedo JP, Salazar O, Andrews BA, Asenjo JA (2008) Cloning and fusion expression of a cold-active lipase from marine Antarctic origin. Enzyme Microb Technol 42:371–377
Phadtare S, Severinov K (2010) RNA remodeling and gene regulation by cold shock proteins. RNA Biol 7:788–795
Phadtare S, Alsina J, Inouye M (1999) Cold-shock response and cold-shock proteins. Curr Opin Microbiol 2:175–180
Priefer UB, Simon R, Puhler A (1985) Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol 163:324–330
Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK (2016) Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24
Przybylak R, Arazny A (2006) Climatic conditions of the north-western part of Oscar II Land (Spitsbergen) in the period between 1975 and 2000. Pol. Polar Res 27:133–152
Repetto M (2012) Lipid peroxidation: chemical mechanism, biological implications and analytical determination. In: Catala A (ed) Lipid peroxidation. InTech, Rijeka, Croatia, pp 3–30
Romaniuk K, Krucon T, Decewicz P, Gorecki A, Dziewit L (2017) Molecular characterization of the pA3J1 plasmid from the psychrotolerant Antarctic bacterium Pseudomonas sp. ANT_J3. Plasmid 92:49–56
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Santiago M, Ramirez-Sarmiento CA, Zamora RA, Parra LP (2016) Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front Microbiol 7:1408
Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Song W, Lin X, Huang X (2012) Characterization and expression analysis of three cold shock protein (CSP) genes under different stress conditions in the Antarctic bacterium Psychrobacter sp. G. Polar Biol 35:1515–1524
Szpirer CY, Faelen M, Couturier M (2001) Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J Bacteriol 183:2101–2110
Touati D (2000) Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6
Tutino ML, Duilio A, Moretti MA, Sannia G, Marino G (2000) A rolling-circle plasmid from Psychrobacter sp. TA144: evidence for a novel rep subfamily. Biochem Biophys Res Commun 274:488–495
Zecchinon L, Claverie P, Collins T, D’Amico S, Delille D, Feller G, Georlette D, Gratia E, Hoyoux A, Meuwis MA, Sonan G, Gerday C (2001) Did psychrophilic enzymes really win the challenge? Extremophiles 5:313–321
Zhang J, Lin S, Zeng R (2007) Cloning, expression, and characterization of a cold-adapted lipase gene from an antarctic deep-sea psychrotrophic bacterium, Psychrobacter sp 7195. J Microbiol Biotechnol 17:604–610
Zheng S, Ponder MA, Shih JY, Tiedje JM, Thomashow MF, Lubman DM (2007) A proteomic analysis of Psychrobacter articus 273-4 adaptation to low temperature and salinity using a 2-D liquid mapping approach. Electrophoresis 28:467–488
Ziegler C, Bremer E, Kramer R (2010) The BCCT family of carriers: from physiology to crystal structure. Mol Microbiol 78:13–34
Acknowledgements
We would like to sincerely thank Antoni Szych for his valuable advice regarding bioinformatic analyses. This work was supported by the Ministry of Science and Higher Education, Poland (Grant number: DI2013 012543) funded from the budget for science in the years 2014–2016, as a research project under the program Diamond Grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ciok, A., Dziewit, L. Exploring the genome of Arctic Psychrobacter sp. DAB_AL32B and construction of novel Psychrobacter-specific cloning vectors of an increased carrying capacity. Arch Microbiol 201, 559–569 (2019). https://doi.org/10.1007/s00203-018-1595-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1595-y