Abstract
The ability to control the transition from an undifferentiated stem cell to a specific cell fate is one of the key techniques that are required for the application of interventional technologies to regenerative medicine and the treatment of tumors and metastases and of neurodegenerative diseases. Reprogramming technologies, which include somatic cell nuclear transfer, induced pluripotent stem cells, and the direct reprogramming of specific cell lineages, have the potential to alter cell plasticity in translational medicine for cancer treatment. The characterization of cancer stem cells (CSCs), the identification of oncogene and tumor suppressor genes for CSCs, and the epigenetic study of CSCs and their microenvironments are important topics. This review summarizes the application of cell reprogramming technologies to cancer modeling and treatment and discusses possible obstacles, such as genetic and epigenetic alterations in cancer cells, as well as the strategies that can be used to overcome these obstacles to cancer research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of human embryonic stem cells (ESCs) is a promising approach in the clinical applications of regenerative medicine and cancer research. However, the use of such ESC derivatives poses a major ethical dilemma, in that embryos need to be destroyed or compromised to produce ESCs. Therefore, more pragmatic alternatives, including reprogramming, are required to pave the path for the clinical application of pluripotent stem cells (PSCs) in humans. Currently, reprogramming technologies are divided into three approaches: (i) somatic cell nuclear transfer (SCNT) technology, (ii) induced PSC (iPSC) technology, and (iii) direct reprogramming (DR) technology [1].
SCNT, iPSC, and DR technologies
SCNT technology generates totipotent cells using an enucleated oocyte injected with a nucleus isolated from differentiated somatic cells [2]. In mammals, the reprogramming capability of somatic cells to an undifferentiated state was first substantiated by the birth of cloned sheep [3]. In a rather different context, ESCs derived from the inner cell mass cells of blastocysts also exhibit pluripotency with indefinite cell division and the ability to differentiate to all three germ layers [4]. The invention of methods for the induction of human iPSCs derived from somatic cells opened a new era of research, as it allowed researchers to derive an almost infinite number of new iPSCs that can be used as a source for autologous cell-based therapy, disease modeling, drug screening, and biomedical engineering [5,6,7,8,9,10,11,12,13]. The current methodologies generally reprogram somatic cells to iPSCs via serial passages in the presence of reprogramming factors (OCT4, SOX2, KLF4, and c-MYC [OSKM], as well as NANOG and LIN28) under adherent culture conditions on a feeder layer or on extracellular matrix (ECM) components [14].
Reprogramming can also be induced by other methods using chemicals that promote the establishment of the core transcription circuitry of stem cells [15,16,17,18]. For example, an over 200-fold increase in reprogramming efficiency was reported for culture media supplemented with antagonists of transforming growth factor beta (TGF-β) signaling and mitogen-activated kinase/extracellular signal-regulated kinase (MEK–ERK) inhibitors, and by passaging the cells in the presence of thiazovivin, which is an inhibitor of the Rho-associated coiled-coil containing protein kinase (ROCK) [19].
A potentially important twist to reprogramming techniques has stemmed from the observation that pluripotency factors, such as OCT4 and LIN28, are markers of a group of stem cell-like cells in ovarian cancers [20]. Several studies have shown that the pluripotency factors used to generate iPSCs also exhibit tumorigenic capability, suggesting that reprogramming and cellular transformation might occur via overlapping pathways [21,22,23,24,25,26,27,28].
Therefore, reprogramming protocols involving the expression of oncogenic pluripotency factors might cause tumorigenesis by disrupting the epigenetic marks for the correct gene expression circuitry. For example, inhibition of the expression of the tumor suppressor gene encoding TP53 not only enhanced the reprogramming of fibroblasts into iPSCs [29], but also generated transformed CSCs from differentiated cells [30]. Moreover, it has been demonstrated that overexpression of c-MYC in immortalized mammary epithelial cells favored tumor formation via epigenetic cell reprogramming [31]. The authors provided evidence that this tumorigenesis was caused by epigenetic reprogramming, as the oncogenic enhancers were reactivated in the cancer cell counterparts. Furthermore, recent works have illustrated an important role of the epigenetic reprogramming of chromatin modifications in the evolution of cancer metastasis [32,33,34]. These articles emphasize the fact that reprogramming can lead to the formation of tumor-initiating cells that acquire stem cell-like phenotypes. Interestingly, the three-dimensional (3D) tumor sphere-forming assay is a unique model of cancer that can be used to investigate malignant heterogeneity in tumorigenesis [34, 35]. Therefore, the tumorigenic potential of the use of reprogrammed stem cells for clinical applications should be recognized and new approaches for safe stem cell therapy should be developed.
DR (or transdifferentiation) technology, which reprograms somatic cells to other differentiated lineages or multipotent stem cells or progenitors, has also been developed [36]. DR introduces target cell-specific, defined transcription factors into recipient somatic cells, which are reprogrammed to the target cells by bypassing the pluripotent stage during lineage conversion, thus possibly avoiding teratoma formation [37,38,39]. Human and mouse somatic cells have been converted into myoblasts, beta islet cells, neurons, and neural stem cells using DR technology [40,41,42,43]. DR is assumed to shorten the preparation period for cell replacement therapy and has the highest potential for clinical application. Nevertheless, to obtain the final target cells, this technique remains time consuming in practice. In addition, compared with SCNT and iPSC technologies, DR exhibits the lowest efficiency of successful reprogramming to PSCs [44, 45]. This problem needs to be overcome.
Cancer stem cells (CSCs)
The CSC hypothesis was proposed over 140 years ago [46] and postulates that cancers arise from a rare subpopulation of cells that are endowed with both tumor and stem cell features. CSCs are resistant to drug and radiation therapies. It is believed that CSCs are self-renewing cancer cells that have clonal tumor-developing capability and clonal long-term repopulation ability [24, 47,48,49]. One model proposes that CSCs are derived from genetically and epigenetically altered stem or progenitor cells that reside in their original niches and acquire oncogenic growth advantages to sustain tumor mass. Thus, these CSCs might possess similar features to normal stem cells and are well adapted to the niche environments [50].
Cancer cell reprogramming
An identical set of reprogramming factors (OSKM) can be delivered to cancer cells derived from almost all tissues to generate induced pluripotent cancer cells (iPCCs) [51,52,53]. Such iPCCs appear to have a CSC-like state after the reprogramming process [49, 53,54,55]. Alternatively, depending on the type of cancer, the introduction of a single gene (a process referred to as DR, see above) can be sufficient to activate multipotency and induce tumor formation. In normally unipotent basal or luminal mouse mammary epithelium, the induction of an activated gene encoding the phosphatidylinositol-4,5-biphosphate kinase catalytic subunit alpha (PIK3CA) was sufficient to trigger the reprogramming of these cells into a multipotent mammary epithelial stem cell-like state and to give rise to breast tumors that displayed a similar cellular heterogeneity to that of human breast cancers [55, 56]. Slightly elevated levels of c-MYC were sufficient to reprogram and dedifferentiate luminal mammary epithelial cells into a stem cell-like state, resulting in the widespread decommissioning of transcriptional enhancers associated with differentiation-specific genes and the reactivation of genes associated with multipotent breast epithelial stem cells [31]. The latter study also showed that c-MYC and an activated PIK3CA allele collaborate in inducing multipotency and in increasing the number of tumor-initiating cells. Schwitalla et al. [32] demonstrated that enhanced NF-kB signaling was able to activate WNT in intestinal cells and induced dedifferentiation of nonstem cells to acquire stem cell-like properties. Another report showed that the constitutively active SMAD2/3 can interact with other factors on OCT4 target loci and potentiate DR conversion with multiple types of transcription factors from myoblasts to adipocytes, B cells to macrophages, and fibroblasts to neurons. Thus, they might be the common cofactors that potentiate diverse cell fate conversions with master genes [57].
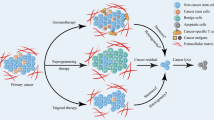
Accordingly, cancer cells that have been reprogrammed via the introduction of a single or several genes, which are capable of triggering a stem cell phenotype, can be a good model of several aspects of cancer research (Fig. 1), such as the study of cancer heterogeneity and niches, the elucidation of the mechanisms of cancer initiation and progression, epigenetic reprogramming, screening of compounds as therapeutic or re-differentiating agents, and induction of cell death/senescence for cancer ablation therapy [58, 59].
Schematic model of the interaction between reprogramming to pluripotency and tumorigenesis. The reprogramming of somatic cells to pluripotency is performed by overexpressing reprogramming factors (such as OCT4, KLF4, SOX2, c-MYC, NANOG, and miRNAs) and inhibiting tumor suppressor genes (such as those encoding p14ARF, p16Ink4a, p21Cip1, and p53), to reset their fate toward a state of pluripotency, which is a dedifferentiation process that resembles tumor development. Patient-specific or healthy iPSCs are used in cell-based therapy after inducing differentiation to appropriate types of cells, for transplantation into patients. For example, iPCCs were derived by introducing OSKM factors and knocking down vector shTP53 in tumor cells in a manner similar to that described in iPSC protocols. The teratomas that are formed after the transfer of iPCCs to SCID mice are then dissected out, and isolated cells can form putative CSC-like phenotypes. The various malignancy characteristics observed in iPCCs seem to depend on differences in tumor cell types. In contrast, CSCs can be derived by an OCT4-mediated dedifferentiation process in tumor progression, even in somatic cells, via the stable expression of telomerase, the H-Ras V12 mutant, and inhibition of the p53 and retinoblastoma protein (pRB) pathways. CSCs can also be derived directly from tumor cells via the overexpression of OCT4, NANOG, KLF4, and IGFBP3, in a dedifferentiating manner. Putative CSCs and iPCCs are expected to be used in studies of drug screening or cancer-initiation mechanisms in the field of human cancer therapeutics. Hypoxia enhances the reprogramming of somatic cells, and HIFs directly regulate the factors that are needed for self-renewal and multipotency in cancer cells and CSCs. Furthermore, hypoxia increases the production of ROS, which promote cell development and EMT in CSCs via the TGF-β signaling pathway and drive CSCs to produce VEGF, which induces angiogenesis
Advantages provided for cancer research by cancer cell reprogramming
Heterogeneity of cancer cells in the same patient can arise for multiple reasons [58,59,60]. First, heterogeneity can be generated by stochastic genetic [61] or epigenetic changes [62]. Clonal evolution confers heritable differences among cancer cells. Second, heterogeneity can arise through the interaction between cancer cells and environmental alterations within the tumors [63]. Third, heterogeneity can be derived from a minor subpopulation of tumorigenic CSCs, which can generate diverse non-tumorigenic cells and constitute a tumor mass. In addition, tumorigenic CSCs can be transplanted between immune-deficient mice and reestablish phenotypic heterogeneity inside the newly formed tumor [64]. These sources of heterogeneity are not mutually exclusive [65]. The cellular heterogeneity encountered in many human tumors may represent a specific niche, which is an important contributing factor to the maintenance of tumor stem cells.
Genetically, some tumors, such as medulloblastomas [66], hepatocellular carcinomas [67], small cell lung carcinomas (SCLCs) [68], and pancreatic adenocarcinomas [69], are composed of such genetic subpopulations of each specific cancer [70]. Different cell subpopulations with a certain cell identity may be dominantly represented by cell reprogramming. For example, breast cancer cells with PIK3CA mutations may present as CSCs after MYC-induced reprogramming [31]. Thus, reprogramming technology might help identify heterologous subsets among cancer progenitor cells. Single-cell RNA sequencing technology has now been used to identify and quantitate these subtypes of cancer cells [71]. Such high-throughput single-cell RNA sequencing technologies have the potential to promote the understanding of cancer generation in the decomposition of heterogeneous cell populations and of the heterogeneity of cells associated with various tumorigenic stages. This technology allows the identification of the cellular subpopulations and the delineation of novel cell markers in the hematopoietic [71], respiratory [72], hepatobiliary [73], and pancreatic [74, 75] lineages, as well as in the intestine [76]. Recent progress in single-cell RNA sequencing led to the identification of the heterogeneous origins of CSCs in gliomas [77], breast cancers [78], myeloid leukemias [79], bladder cancers [80], and colorectal cancers [81]. These heterogeneities stem from the original unidentified subpopulations that arise during the step of induction of cancer-specific iPSCs. Henceforth, this single-cell sequencing technique can be applied to cancer-specific iPSCs. The limitations of this technique in the identification of heterogeneity are also presented. The major sources of genetic and phenotypic variations among iPSCs can be assessed by including gene copy numbers and the extent of epigenetic changes, indicating that this problem could be solved in the near future and that the real heterogeneity of such CSCs may be identified [82].
Study of the microenvironmental niches of cancer stemness and organoid culture
Patient-specific iPSCs could be applied as cancer models for mechanistic studies and drug development, as well as for studying interactions with cancer niches. The newly developed 3D cell cultures of patient-derived iPSCs might be useful for understanding the roles of cellular microenvironments. Primary tumors at a late stage of cancer development represent differentiated cell lineages. The animal models that are available currently fail to provide ideal systems because of their genetic differences. The 3D and organoid cell culture of iPSCs could provide useful systems that are appropriate for modeling human cancers, which would be clinically important for drug screening and the development of therapies.
Stem cells grow in their own cellular microenvironments, termed stem cell niches. In these niches, both interactions between cells and the ECM and their diffusible signals are important for development. Niches have been used to identify mammalian stem cells in various epithelial tissues from normal samples and cancers [83]. The niches are composed of fibroblasts, immune cells, endothelial and vascular progenitor cells, or ECMs and network signals composed of cytokines and growth factors [79]. CSCs are also capable of forming niches representing tumor microenvironments [84]. During tumor progression to a more malignant stage, CSCs in the primary tumor depend on the tumor microenvironment or on the CSC niches that are located within it [85]. Thus, the reprogramming of cancer cells to generate iPCCs can provide critical information that can be used to understand the role of such microenvironments.
To elucidate the role of niches, the recently developed technique of organogenesis also provides useful information [86]. Recent techniques of organoid formation from brain, intestine, kidney, liver, lung, ovary, pancreas, and stomach cells provide basic knowledge on the cross-talk between CSCs and their microenvironment. This technique can be used in clinical applications, including cancer modeling, drug screening, microorganism infection, and therapy using new gene editing technologies, such as CRISPR/Cas9, to identify the critical genes, respectively. Thus, patient-derived organoids might be critical for future use in cancer research, for drug screening, and for mechanistic studies of CSCs and their microenvironments [87].
Merits of the application of this technique to cancer modeling and cancer therapy
Cancer cell reprogramming can be used as a model to understand tumorigenesis and to develop regenerative therapies. In some cases, such reprogramming advances oncogenic capacity even further. Thus, after dedifferentiation, reprogrammed cancer cells exhibit a more severe cancer phenotype because of the genetic alterations or oncogenicity of the reprogramming factors that were used [40, 52, 88,89,90].
Leukemia
The reprogramming of the chronic leukemia KBM7 line into iPCCs using the transcription factors OSKM led to resistance to an inhibitor of the Bcl–Abl fusion oncogene in these cells, but not in the parental cells [52]. In another case, primary chronic myelogenous leukemia (CML)-derived iPCCs were shown to be resistant to imatinib. However, CML-iPCCs-derived hematopoietic cells recovered sensitivity to this drug. These findings indicate that the pathological features of the initial disease were recapitulated [88].
Gastrointestinal cancers
Nagai et al. [90] also reprogrammed gastrointestinal cancer cell (GCC) lines using OSKM. These iPCCs were sensitized to chemotherapeutic drugs and differentiation-inducing protocols at an early stage, but longer culture of these cells resulted in more aggressive features compared with the parental cells. Thus, the authors speculated that the cancer-specific iPCCs were prone to genetic instability via genetic or epigenetic alterations, including oncogenic c-Myc activation. Human pancreatic ductal adenocarcinoma (PDAC) cells were reprogrammed to generate iPCCs and injected into SCID mice. The reprogrammed cancer cells then produced the pancreatic intra-epithelial neoplastic lesions that can progress to invasive tumors [40]. Miyoshi et al. [53] used four different GCC lines to obtain iPSC-like cells. These GCC-iPSCs were generated by ectopic expression of OSKM and oncogenes, such as BCL2 and KRAS, and short-hairpin RNAs (shRNAs) against the tumor suppressor genes, such as TP53, p16Ink4a, PTEN, FHIT, or RB1. These iPSC-like cells were more sensitive to 5-fluorouracil and drugs of differentiation–induction and exhibited reduced tumorigenicity in nonobese diabetic/severe combined immunodeficient mice. Kuo et al. [58] found that the positive feedback between OCT4 and c-JUN increased with the onset of cancers. We hypothesized that the positive feedback regulation of OCT4 and c-JUN might promote the generation of liver CSCs.
Lung cancers
Mahalingam et al. [91] reprogrammed a non-small cell lung cancer (NSCLC) cell line using OSKM to generate NSCLC-iPCCs, which reversed the aberrantly dysregulated genes in cancer cells both epigenetically and transcriptionally, resulting in reduced oncogenicity in iPCCs.
Li‒Fraumeni syndrome (LFS)
LFS is a cancer hereditary syndrome caused by TP53 germline mutations. Patients with LFS are susceptible to adrenocortical carcinoma, brain tumor, breast cancer, leukemia, osteosarcoma, and soft tissue sarcoma. LFS-patient-derived iPSCs have been generated [92]. LFS-iPSC-derived osteoblasts reproduced the hallmarks of osteosarcoma (OS), including defective osteoblastic differentiation and tumorigenicity. However, osteoblasts from LFS-derived iPSCs did not exhibit cytogenetic alterations in 18 regions that are usually associated with late-stage OS. The imprinting gene H19 was not upregulated in LFS osteoblasts during osteogenesis, and the restored forced expression of H19 in LFS osteoblasts improved osteoblastic differentiation and suppressed tumorigenicity. Thus, without differentiation, iPSCs were able to maintain stemness with higher expression of the H19 gene product, even though the TP53 gene was mutated.
LFS-derived iPSCs provide several advantages compared with other models of LFS, such as (i) an unlimited supply of cells, (ii) a human platform, and (iii) access to the heterogeneity across cell types. Thus, LFS-derived iPSCs can provide great value in drug screening and testing in vitro. LFS-derived iPSC models enable the understanding of precise genome editing, three-dimensional (3D) organoid-based culturing systems, and subsequent organ-on-chip systems, which might facilitate anticancer drug discovery and provide a sophisticated model of cancer treatment [92].
Merits of the development of therapeutics
A cell line of the blast crisis stage of CML was reprogrammed to generate CML-iPSCs [52]. CML was generated by mutating the BCR–ABL fusion gene, which caused enhanced cell expansion [93], while CML-iPSCs retained their differentiation potential. Thus, the maintenance of stemness and oncogenic expansion is a critical issue during differentiation. In a blast crisis, cells lose their ability to differentiate, and immature leukemia cells can overgrow instead. In the case of in vivo differentiation in teratomas, CML-iPSCs differentiate into all three germ layers, including hematopoietic cell lineages expressing CD34, CD43, and CD45. Cells with loss of the CML phenotype and independence from BCR–ABL signaling were resistant to imatinib. Differentiation of the cells into hematopoietic lineages in vitro rendered them sensitive to imatinib, suggesting the recovery of oncogenic dependency, as the CML-iPSCs underwent hematopoietic differentiation.
Kumano et al. [88] demonstrated that iPSCs derived from the primary tumors of two patients with CML exhibited stemness and differentiation to hematopoietic progenitors that expressed BCR–ABL. These iPSCs were prepared from imatinib-sensitive patients, but the iPSCs finally showed resistance to this drug and resembled CML stem cells after reprogramming. These cell lines might provide a good model system for understanding the mechanism of drug resistance and the role of stem cells in CML.
iPSCs might be useful for the development of personalized approaches to cancer treatment, as they would enable the discovery of a wide range of therapeutic agents against the genetic differences between individuals, which might aid the discovery of those that are ideal for each patient [94]. The identification of an efficient strategy to eliminate CSCs is a critical issue in cancer therapy. As CSCs are rare, iPSC technologies could be used to generate a large quantity of CSCs for subsequent applications [95, 96]. Nishi et al. [97, 98] generated mammary CSC-like cells that were used to screen compounds that selectively targeted CSCs, including salinomycin and withaferin A. Choi et al. [99] generated iPSC-derived hepatic cells from patients with α-1 antitrypsin (AAT) deficiency, to screen the Johns Hopkins Drug Library (3131 clinical compounds). Of the 262 compounds that led to decreased AAT accumulation by > 50%, 43 showed no side effects. Finally, the authors identified five hits that consistently decreased AAT levels in four AAT-deficient iPSC lines. Patient-derived iPSCs are also useful for the study of drug absorption, distribution, metabolism, excretion, and toxicity. Thus, the use of iPSCs is beneficial for the identification of CSC-related genes and for mechanistic studies of cancer induction, promotion, and progression.
Study of metabolic shifts
Cancer cell reprogramming has the advantages of reconstituting cancer initiation and progression, which renders it an ideal model to investigate changes in cancer characteristics, such as metabolism, epithelial–mesenchymal transition (EMT)/mesenchymal–epithelial transition (MET), and metastasis.
The Warburg effect, via which cancer cells use glycolysis rather than oxidative phosphorylation in mitochondria for producing energy, is well known [100, 101]. Aerobic glycolysis, which is mediated by uncoupling proteins that uncouple oxidative phosphorylation from glycolysis [102,103,104,105], is enhanced in ovarian and breast cancers and when PSC pluripotency is induced. Lu et al. [106] generated iPSCs from patients with ataxia telangiectasia (AT) syndrome that mimicked the AT phenotype, including deregulated AT-mutated (ATM)-associated pathways and altered gene expression patterns in the pentose phosphate and mitochondrial oxidative phosphorylation pathways. Metabolic reprogramming of pyruvate utilization is a therapeutic target for the development of new reagents for cancer prevention [107], such as those affecting the inhibition of pyruvate dehydrogenase kinase [108]. The anti-hyperglycemic agent metformin is an interesting substance with therapeutic effectiveness. Although the action of metformin has not been explained fully, it is useful for the metabolic reprogramming of cancer cells [109]. Metformin promoted growth arrest in pancreatic tumor cells via direct impairment of fatty acid synthesis [110]. The antitumor effects of metformin appear to be correlated with microRNA (miRNA) modulation and increased expression of the AMP-activated protein kinase, leading to the modulation of targets that restore energy homeostasis by inhibiting hepatic gluconeogenesis [109].
Analysis of EMT/MET
EMT/MET play critical roles during normal development, as they contribute to the formation of the mesoderm during gastrulation, as well as at subsequent stages of the development of neural crests and lung formation [111]. They are also hallmark of cancer initiation and metastasis. For example, EMT/MET inducers, such as SNAIL1/2 or TWIST1/2, are associated with relapse and survival in several cancers, such as those that arise in mammary, colorectal, and ovarian tissues, suggesting that EMT/MET pathways are associated with poor outcomes of cancer patients [112, 113]. The expression of EMT/MET genes is correlated with cancer progression in colon cancers, papillary thyroid carcinomas, and breast carcinomas [113], and in the development of metastases in melanomas [114]. In xenotransplantation assays, iPSCs derived from human sarcoma cell lines proliferated more slowly than did their parental counterparts and exhibited necrosis and lower expression of EMT markers [115]. During reprogramming, initial methylation followed by demethylation of the promoters of 32 oncogenes and 82 tumor suppressor genes were demonstrated, showing that pluripotency factors can suppress the features of cancer phenotypes, restore differentiation potentials, perturb epigenetics via DNA methylation, and alter cancer-related gene expression.
Molecular approach to the study of cancer metastasis
Compared with their normal counterparts, cancer cells exhibit widespread alterations in DNA methylation patterns and an altered organization of open and condensed chromatin because of profound changes in epigenetic chromatin marks [116, 117]. Additional epigenomic reorganization takes place during tumor progression to metastasis [118, 119]. Each metastatic event establishes a new tumor nodule and is, thus, by definition, carried out by CSCs [120]. Recent studies have begun to shed light on the molecular mechanisms that lead to metastasis. Not surprisingly, the changes appear to be tumor-type specific. For example, during SCLC progression to metastasis, the expression of the transcription factor nuclear factor 1b (Nfib) increases by several fold, in part from the amplification of the Nfib gene, resulting in the activation of new distal regulatory elements (i.e., transcriptional enhancers) and the implementation of a neuroendocrine transcriptional program that drives metastasis [121]. In PDAC, the genomes of primary tumors and their metastases are largely similar, suggesting that epigenetic reprogramming might be the primary force driving the transition [122]. Two different reports have described widespread chromatin and gene-enhancer reprogramming during PDAC progression [33]. Those authors investigated matched PDAC cells from the same patients from either proximal (peritoneum) or distant (lung and liver) metastatic sites. PDAC metastases from distant sites were dependent on the oxidative pentose phosphate pathway for the maintenance of their malignant gene expression programs. Roe et al. [34] also used a mouse PDAC model and found that the transition to a metastatic state was accompanied by massive FoxA1-driven enhancer activation. The newly activated genes rendered cells more invasive, and they assumed a cell fate resembling that of the embryonic foregut endoderm.
These examples suggest that the reprogrammed cancer cells displayed various cancer phenotypes that provided a prevention technology and insights into cancer biology and the progression of cancers.
Obstacles to cancer cell reprogramming
This reprogramming technique for cancer cells remains immature; therefore, additional trials are needed to understand the weakness that exists currently in cell reprogramming for the translational research of cancers.
Mutations in the genome
Usually, cancers are produced by “driver” mutations at the initiation stage and, subsequently, by positive selection and clonal expansion, which lead to the accumulation of “passenger” mutations [123, 124]. The “driver” mutations confer an advantage to the proliferation and development of cancers. In contrast, the “passenger” mutations do not affect the fitness of cancer clones significantly [125,126,127]. Recent advances in deep genomic sequencing technologies have led to the identification of these mutations in some oncogenes and tumor suppressor genes, which are the hallmark drivers of certain cancers [128]. However, whether these genetic mutations become a barrier to cancer cell reprogramming remains unclear.
In addition, many studies have demonstrated that the process of cell reprogramming may cause genomic alterations, such as chromosomal aberrations, copy number variations (CNVs), and single-nucleotide variations. For example, trisomy 12 is an aberration that is observed commonly in ESCs and iPSCs [72,73,74,75]. Some cell cycle-related genes and NANOG are located on chromosome 12; thus, trisomy 12 might result in alterations in proliferation and reprogramming [76, 77]. The amplification of chromosomes 8 and X, as well as of other chromosomes, was also detected in iPSCs [72, 73]. iPSCs may acquire CNVs during reprogramming or from the mosaicism that is present in the parental cells; however, CNVs are lost gradually by cell passaging, with selective pressure for the deletion of tumor suppressor genes in early cell passages and duplication of oncogenes at a later time [75, 82,83,84, 129]. Single-nucleotide mutants in iPSCs are identified by high-throughput next-generation sequencing analyses. These analyses have identified an average of ten protein-coding mutations per human iPSC line [68, 85]. Thus, further investigation is required to identify approaches aimed at preventing these mutations during the cellular reprogramming of cancer cells.
The use of young donor cells is one possible way to overcome this issue, because mutations in mitochondrial DNA increase with age in human iPSCs [88, 130]. If the preparation of autologous donor cells is difficult, human histocompatibility antigen (HLA)-matched allogenic cells can be used to replace them in reprogramming, to generate iPSCs. It might not be necessary to prepare autologous donor cells, because human HLA-matched umbilical cord blood-derived iPSCs, which do not show a higher rate of point mutations, are useful sources of allogenic iPSC-based cell therapies [131]. Yamanaka’s group and the RIKEN Cell Bank in Japan are initiating this project to cover most Japanese HLAs to produce allogenic iPSCs with lower mutation rates that could be used as iPSCs bank stocks.
Epigenetic alterations
The process of fibroblast reprogramming using Yamanaka’s factors (OSKM) includes three steps: initiation, maturation, and stabilization [132]. The initiation step is characterized by the expression of genes that encode proteins involved in MET via the silencing of SNAIL1/2, suppression of TGF-β signaling, and upregulation of CDH1 [133]. In the maturation step, the expression of exogenous 4Fs is repressed and pluripotent-related genes, such as NANOG, SALL4, and ESRRB, are expressed in their stead. In the stabilization step, other pluripotent marker genes are expressed for full reprogramming.
In the initiation step, cells undergoing reprogramming exhibit downregulation of the H3K79me2 epigenetic markers located around MET-related genes. A decreased H3K79me2 level indicates the inhibition of mesenchymal properties through transcriptional repression. Subsequently, the genes encoding poly-(ADP-ribose) polymerase-1 and the ten–eleven translocation (TET) family 2 (TET2) are recruited to the NANOG and ESRRB loci, which direct the transition from the initiation to the maturation phase [134]. In the maturation and stabilization phases, epigenetic silencing of the exogenous genes and enhancing of chromatin remodeling represent the resetting of epigenetic modifications in these reprogramming-related genes [135].
TET-mediated DNA demethylation at CpG islands (at the ESRRB and OCT4 loci via an interaction with NANOG) promotes gene expression and helps maintain the pluripotency of stem cells [136]. In cancer cells, high levels of DNA methyltransferase 1 (DNMT1) and DNA methyltransferase 3A/3B (DNMT3A/3B), as well as suppression of TET, have been detected [137, 138]. The repressed function of TET in cancer cells might impair pluripotency and genomic reprogramming. The epigenetic features of cancer cells, such as high expression of DNMTs, low expression of TETs, and overexpression of histone deacetylases (HDACs), might be an obstacle to the reprogramming process.
In addition to DNA methylation, histone modifications also play critical roles in cancer cell reprogramming (Fig. 2). Histone marks, such as H3K27me3, H3K9me3, H3K4me3, and H3K27ac, are targets for the reprogramming of cancer cells. The catalytic subunit of the polycomb repressive complex 2 (PRC2) enhancer of zesta homolog2 (EZH2) mediated transcriptional repression by introducing H3K27me3 [139]. In breast cancers, B-cell lymphomas, and prostate cancer, EZH2-mediated H3K27me3 permitted the silencing of tumor suppressor genes [140,141,142,143]. Accordingly, in myeloid malignancies, loss of EZH2 function was sufficient to induce a self-renewal-supporting transcriptional program and leukemogenesis. These reports indicate that the deregulation of the H3K27me3 landscape—hence, the transcriptional repression—is the driving force behind the emergence of CSCs, independent of the original EZH2 mutation [144,145,146,147]. The mixed lineage leukemia (MLL) histone methyltransferase is also involved in histone modification. MLL requires the repressive activity of the polycomb repression complex 1 (PRC1), which monoubiquitinates histone H2A at lysine 119 (H2AK119Ub1) or trimethylates histone H3 at lysine 4 (H3K4me3), and then cooperates with PRC2 to mediate transcriptional repression [139]. The Bmi1 subunit of PRC1 mediates the repression of tumor suppressors in myeloid progenitors [148, 149] and is required for the inhibition of tumor suppressor genes that is necessary to initiate the self-renewal of CSCs in solid tumors [150]. The control afforded by ATP-dependent chromatin remodeling complexes, such as SWI/SNF, ISWI, CHD, and INO080, represents another pathway of epigenetic regulation in mammals [151]. The genes encoding the SWI/SNF complex are mutated in > 20% of human cancers. Loss of SMARCB1, a subunit of the SWI/SNF complex, drives malignant rhabdoid tumors and is associated with the blocking of differentiation, reprogramming toward an oncogenic transcriptional program, and activation of cancer signaling [152, 153]. ARID1A, another subunit of SWI/SNF, is a tumor suppressor in colon cancers and its loss activates an oncogenic program and promotes the development of invasive colon adenocarcinomas in the mouse [154]. Taken together, these findings show that deregulations of the DNA methylation and histone modification landscapes represent key steps in the onset of the generation of CSCs.
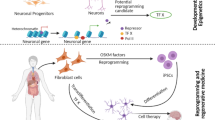
Schematic model of the mechanisms via which epigenesis, p53, and ROS‒hypoxia‒HIFs promote reprogramming efficiently and genome integrity in PSCs. Cancer cells with driver and passenger mutations might be overcome by epigenetic reprogramming and DNA repair to induce the formation of PSCs with correct plasticity. Active chromatin with active histone markers (H3K4me3, H3K79me2, H3Ac, and H3K27Ac) should be repressed by repressive markers (H3K9me3, H3K36me2/3, and H3K27me3) at specific regions by three different reprogramming methods (SCNT, iPSC, and DR). Forced expression of reprogramming factors increases the levels of ROS that are generated in mitochondria, which in turn causes DNA damage and undermines both reprogramming efficiency and the genomic integrity of iPSCs. Antioxidants can promote reprogramming efficiency and safeguard the stability of the genomes of iPSCs by inhibiting ROS production and exerting non-antioxidant functions, including modulating epigenetic modifiers, and histones
In general, it might be better to define precise chromatin regulatory regions, including physical constraints such as the insulators and topologically associated domains, while lamina-associated domains are mainly localized along large organized chromatin modifications, as well as with the heterochromatic regions of silenced genes in cells [155]. Alterations in these higher order structures have been linked to the control of tumorigenesis [156]. Transcriptional enhancers are enriched for the binding of chromatin factors such as p300/CBP, a major histone acetyltransferase that mediates the formation of H3K27AC, and its mediator, a long-range interaction facilitator [157]. The correct ordering and functional integrity of these modifiers with transcription factors and enhancers should be clarified in terms of the generation and expansion of CSCs [158].
However, recent development in high-throughput tools that allow the examination of chromatin structure, such as DNase I-, formaldehyde-assisted isolation of regulatory elements- (FAIRE-), and assay for transposase-accessible chromatin (ATAC) sequencing, can be used to extend our knowledge of epigenetic regulation during cell reprogramming [159, 160]. Conversely, the incorporation of hyperdynamic histone variants at enhancers (H2A.Z and H3.3) might render the chromatin less stable and facilitate the initial access to transcription factors [161,162,163], demonstrating the presence of oncogenic enhancers that are involved in cancer commitments. However, many questions remain unanswered: how can cancer reprogramming erase the epigenetic memory of stem or differentiated cells? How can oncogenic enhancers be maintained? Which molecules are involved in the initiation and progression of cancer genotypes with expanding CSCs of each tumor type [164]? CRISPR-mediated epigenome editing may be a promising technique to identify the key cis-elements in the genomes of CSCs [165,166,167,168]. The origins of cancer stemness and the manners in which stemness genes and oncogenes might be separated remain unclear. However, these breakthroughs and the identification of new drugs targeting epigenetic processes (epi-drugs) may open a new era of therapeutic strategies to target CSCs for reprogramming.
Potential key factors to overcome the obstacles to cancer cell reprogramming
Several reprogramming enhancers are thought to be able to overcome the problems raised above. These reprogramming enhancers can be divided into the following categories: modulators of tumor suppressor proteins, hypoxia and reactive oxygen species (ROS), and cellular signaling and chromatin modifiers (Table 1, Fig. 2).
Tumor suppressor proteins
Transient inhibition of the gene encoding the tumor suppressor protein 53 (TP53) or the phosphatase and tensin homolog protein (PTEN) increases reprogramming efficiency [51, 169,170,171,172,173,174,175]. During the transient inhibition of tumor suppressors, cell proliferation is increased, and cell cycle arrest, apoptosis, and senescence are inhibited, which are favorable conditions for reprogramming. For example, the introduction of a dominant-negative TP53 [176] or shRNA–TP53 [59] into cells increased the efficiency of reprogramming. However, cyclin D1 was reported to be an obstacle to reprogramming to a pluripotent state [177]. Inhibition of TP53 was also effective in the direct conversion of human fibroblasts to dopaminergic neurons [178].
Hypoxia and ROS scavenger JDP2
Hypoxia induces the expression of hypoxia-inducible factors (HIFs). Two main HIFs, HIF1α and HIF2α, are essential for the metabolic changes that are required to generate iPSCs, whereas HIF2α is detrimental at the late stage of reprogramming of human cells. Prolonged HIF2α stabilization represses reprogramming because the tumor necrosis factor (TNF)-related apoptosis-inducing ligand and apoptosis are induced. Hypoxia treatment used during the induction of iPCCs might induce an increase in tumorigenicity, indicating the possibility that the targets of HIFs might be enhancers of CSC genes [89]. Moreover, hypoxia and expression of HIFs are required for the survival of CSCs [47, 179] and trigger ROS-dependent EMT [180]. Both hypoxia and elevated levels of glycolysis are conducive to the maintenance of stem cell features. It has been proposed that hypoxic culture conditions and reduced mitochondrial respiratory activity might increase the generation of iPSCs and inhibit the differentiation of ESCs [181, 182]. For example, it has been shown that hypoxia increases the DR efficiency of somatic cells into induced neural stem cells (iNSCs) or induced cardiomyocytes (iCMs) [183].
ROS are toxic oxygen derivatives and radicals derived from aerobic metabolism that lead to cellular damage and cell death [184, 185]. Increased levels of ROS reduce cell viability and decrease the reprogramming efficiency. In contrast, ROS scavengers lower oxidative stress, thereby increasing reprogramming efficiency. The reprogramming efficiency is significantly increased by adding vitamin C to the cell reprogramming culture medium [186]. The c-Jun dimerization protein 2 (JDP2) was identified as a cofactor that enhances antioxidant response activity [187, 188]. JDP2 acts as a repressor protein that inhibits cell proliferation; it induces cellular senescence during tumor development and participates in ROS homeostasis to inhibit cell damage by ROS [188]. These molecular features of JDP2 are also controlled by hypoxia and HIFs. Oxidative stress also induces angiogenesis via increased expression of angiogenetic marker genes, such as the vascular endothelial growth factor (VEGF) gene [189]; moreover, hypoxia stimulates the production of VEGF by CSCs [190]. Taken together, these findings suggest that the stemness of CSCs might be affected by extrinsic factors, such as hypoxia, ROS, and signaling between CSCs and environmental niches (e.g., TGF-β and the tumor necrosis factor-α, WNT, NOTCH, SHH signals and ECM stiffness, and some CSC-related transcription factors) [180, 190,191,192,193].
Signaling modulators and chromatin modifiers
In addition to the reprogramming factors mentioned above, other reprogramming enhancers, including miRNAs and lncRNAs [194], have been emerging. These are also factors that are key to overcoming the obstacles to cancer cell reprogramming (Table 1). Kaufhold et al. [195] found that Yin Yang 1 (YY1) was a transcriptional repressor for stemness factors such as BMI1, SOX2, and OCT4. YY1 contributes to enhancer‒promoter interactions in a manner that is analogous to the DNA interaction mediated by CTCF [196]. The existence of a regulatory loop between the nuclear factor kappa b (NF-kB)–PI3K–AKT pathway and downstream products, such as BMI1, OCT4, SOX2, and YY1, has also been noted. Thus, modulation of YY1 and NF-kB–PI3K–AKT signaling may contribute to cell reprogramming. TGF-β pathway inhibitors, such as SB431542, increased the efficiency of the reprogramming of adult cardiac fibroblasts to iCMs [197]. Moreover, the B-lymphoma Mo-MLV insertion region 1 homolog (Bmi1) is a barrier to cardiac reprogramming. The inhibition of Bmi1 leads to an increase in the level of the active histone mark H3K4me3, and to a decrease in the level of the repressive mark H2AK119ub at cardiogenic loci, resulting in cardiac gene expression and increased reprogramming efficiency [198]. The zinc finger protein 281 (ZNF281) also enhances the direct conversion of fibroblasts to iCMs [199]. Because the high-mobility group AT-hook 2 (HMGA2) is involved in higher order chromatin compaction, its overexpression might help relax the nucleosome into a more open state for DR. Among the known reprogramming factors, the SOX family members, especially those of the SOXB, SOXE, and SOXF subclasses, are potent drivers of direct somatic cell reprogramming into multiple lineages [200].
Chromatin modifiers, such as the EZH2 and ASCL1 components, are also useful for DR to iNSCs or motor neurons [183, 201,202,203]. JMJD3 has been reported as an epigenetic enhancer of lineage conversion from bone marrow progenitors to liver cells [204]. Agathocleous et al. [205] and Cimmino et al. [206] reported that vitamin C regulated HSCs and suppressed leukemogenesis by modulating TET2 activity. Vitamin C is a cofactor of Fe2+- and alpha-ketoglutarate-dependent dioxygenases. Vitamin C modulates stem cell function, potentiates the reprogramming of fibroblasts to iPSCs, and inhibits the aberrant self-renewal of HSCs by enhancing the activity of Jumonji-1C domain-containing histone demethylases or TET DNA hydroxylases. Thus, vitamin C restores TET function in HSCs and might represent an adjuvant agent for treating leukemia and other cancers [207]. Vitamin C treatment has been applied to cancer cells such as melanomas [208], in which it increased 5-hydroxymethylcytosine content and resulted in the inhibition of tumor cell invasion and clonogenic growth in soft agar. Moreover, vitamin C is also useful for the metabolic reprogramming of cancer cells [207]. In addition to these identified reprogramming enhancers/modulators, further studies are still required to verify and overcome the problems of epigenetic reprogramming, mutations, and ROS-metabolic reprogramming in mitochondria and the endoplasmic reticulum.
Conclusions and future perspectives
The basic techniques of cell reprogramming have their own merits for each cell type. Both the SCNT and iPSC technologies have the potential to erase genetic and epigenetic modifications in cancers and return the cells back to their stemness phenotype. Although DR- and classical iPSC-based reprogramming have considerable potential, their low efficiency of successful reprogramming and poor reproducibility limit the development of research in this field. Several obstacles must be overcome in the use of cell reprogramming. It will be challenging to maintain homeostasis, regulate ROS production, and maintain normal aging in the directly reprogrammed and pluripotent cells. Reprogramming enhancers are possible modulators of cancer and their microenvironments (niches) that might allow the application of this technology to translational research. Among the tumor suppressor genes, the status of TP53 signaling in CSCs plays a critical role in maintaining the stemness and expansion of cancer cells [49, 209]. To study therapeutic models in cancer research, 3D organoid models of ductal pancreatic cancers have provided a new spectrum of models of tumor progression by forming neoplasms that proceed to form invasive and metastatic carcinomas [210]. The organoid methodology is a useful system that can be used to identify the characteristics of malignancy, and the creation of complete tissues or neoplastic cancer organoids in vitro might provide better models of cancers in the future [35, 211,212,213,214]. Moreover, the CRISPR/Cas9 approach is believed to be a new breakthrough technology that can be used to correct cancer genomes for clinical applications [214]. However, Haapaniemi et al. [212] and Ihry et al. [213] demonstrated that CRISPR/Cas9 genome editing technology induces p53-mediated DNA damage and that, in human PSCs, p53 inhibits CRISPR/Cas9-induced genome editing. Thus, the tumor suppressor product of TP53 remains critical for overcoming this problem. Efforts to harness the versatility of iPSCs to model human cancers and to screen for effective therapeutics will undoubtedly accelerate translational cancer research from the laboratory to the bedside.
References
Ma T, Xie M, Laurent T, Ding S (2013) Progress in the reprogramming of somatic cells. Circ Res 112:562–574. https://doi.org/10.1161/CIRCRESAHA.111.249235
Gurdon JB (1962) The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol 10:622–640
Campbell KH, McWhir J, Ritchie WA, Wilmut I (1996) Sheep cloned by nuclear transfer from a cultured cell line. Nature 380:64–66. https://doi.org/10.1038/380064a0
Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676. https://doi.org/10.1016/j.cell.2006.07.024
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. https://doi.org/10.1016/j.cell.2007.11.019
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920. https://doi.org/10.1126/science.1151526
Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R (2007) Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318:1920–1923. https://doi.org/10.1126/science.1152092
Okita K, Ichisaka T, Yamanaka S (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317. https://doi.org/10.1038/nature05934
Ebert AD, Yu J, Rose FF Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN (2009) Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457:277–280. https://doi.org/10.1038/nature07677
Saha K, Jaenisch R (2009) Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell 5:584–595. https://doi.org/10.1016/j.stem.2009.11.009
Wu Z, Chen J, Ren J, Bao L, Liao J, Cui C, Rao L, Li H, Gu Y, Dai H, Zhu H, Teng X, Cheng L, Xiao L (2009) Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol 1:46–54. https://doi.org/10.1093/jmcb/mjp003
Wang SW, Wang SS, Wu DC, Lin YC, Ku CC, Wu CC, Chai CY, Lee JN, Tsai EM, Lin CL, Yang RC, Ko YC, Yu HS, Huo C, Chuu CP, Murayama Y, Nakamura Y, Hashimoto S, Matsushima K, Jin C, Eckner R, Lin CS, Saito S, Yokoyama KK (2013) Androgen receptor-mediated apoptosis in bovine testicular induced pluripotent stem cells in response to phthalate esters. Cell Death Dis 4:e907. https://doi.org/10.1038/cddis.2013.420
Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K (2008) A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3:340–345. https://doi.org/10.1016/j.stem.2008.08.003
Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA (2008) Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol 26:795–797. https://doi.org/10.1038/nbt1418
Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K (2009) A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5:491–503. https://doi.org/10.1016/j.stem.2009.09.012
Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S (2009) Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell 4:16–19. https://doi.org/10.1016/j.stem.2008.11.014
Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H (2013) Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341:651–654. https://doi.org/10.1126/science.1239278
Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, Ding S (2009) A chemical platform for improved induction of human iPSCs. Nat Methods 6:805–808. https://doi.org/10.1038/nmeth.1393
Peng S, Maihle NJ, Huang Y (2010) Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 29:2153–2159. https://doi.org/10.1038/onc.2009.500
Almstrup K, Hoei-Hansen CE, Wirkner U, Blake J, Schwager C, Ansorge W, Nielsen JE, Skakkebaek NE, Rajpert-De Meyts E, Leffers H (2004) Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res 64:4736–4743. https://doi.org/10.1158/0008-5472.CAN-04-0679
Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, Brohee S, Salmon I, Dubois C, del Marmol V, Fuks F, Beck B, Blanpain C (2014) SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511:246–250. https://doi.org/10.1038/nature13305
Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y (2008) The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem 283:17969–17978. https://doi.org/10.1074/jbc.M802917200
Clark AT (2007) The stem cell identity of testicular cancer. Stem Cell Rev 3:49–59
Hart AH, Hartley L, Parker K, Ibrahim M, Looijenga LH, Pauchnik M, Chow CW, Robb L (2005) The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer 104:2092–2098. https://doi.org/10.1002/cncr.21435
Hochedlinger K, Yamada Y, Beard C, Jaenisch R (2005) Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121:465–477. https://doi.org/10.1016/j.cell.2005.02.018
Rowland BD, Peeper DS (2006) KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer 6:11–23. https://doi.org/10.1038/nrc1780
West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ (2009) A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460:909–913. https://doi.org/10.1038/nature08210
Krizhanovsky V, Lowe SW (2009) Stem cells: the promises and perils of p53. Nature 460:1085–1086. https://doi.org/10.1038/4601085a
Scaffidi P, Misteli T (2011) In vitro generation of human cells with cancer stem cell properties. Nat Cell Biol 13:1051–1061. https://doi.org/10.1038/ncb2308
Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, Miluzio A, Gaudioso G, Vaira V, Turdo A, Giaggianesi M, Chinnici A, Lipari E, Bicciato S, Bosari S, Todaro M, Zippo A (2018) MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun 9:1024. https://doi.org/10.1038/s41467-018-03264-2
Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, Rupec RA, Gerhard M, Schmid R, Barker N, Clevers H, Lang R, Neumann J, Kirchner T, Taketo MM, van den Brink GR, Sansom OJ, Arkan MC, Greten FR (2013) Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152:25–38. https://doi.org/10.1016/j.cell.2012.12.012
McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, Stauffer KM, Makohon-Moore A, Zhong Y, Wu H, Wellen KE, Locasale JW, Iacobuzio-Donahue CA, Feinberg AP (2017) Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet 49:367–376. https://doi.org/10.1038/ng.3753
Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K, Filippini D, Creighton B, Burkhart RA, Buscaglia JM, Kim EJ, Grem JL, Lazenby AJ, Grunkemeyer JA, Hollingsworth MA, Grandgenett PM, Egeblad M, Park Y, Tuveson DA, Vakoc CR (2017) Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170(875–888):e820. https://doi.org/10.1016/j.cell.2017.07.007
Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Ohlund D, Handly-Santana A, Ardito-Abraham CM, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov GN, Delcuze B, Creighton B, Wright K, Park Y, Morsink FH, Molenaar IQ, Borel Rinkes IH, Cuppen E, Hao Y, Jin Y, Nijman IJ, Iacobuzio-Donahue C, Leach SD, Pappin DJ, Hammell M, Klimstra DS, Basturk O, Hruban RH, Offerhaus GJ, Vries RG, Clevers H, Tuveson DA (2015) Organoid models of human and mouse ductal pancreatic cancer. Cell 160:324–338. https://doi.org/10.1016/j.cell.2014.12.021
Davis RL, Weintraub H, Lassar AB (1987) Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987–1000
Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463:1035–1041. https://doi.org/10.1038/nature08797
Vierbuchen T, Wernig M (2011) Direct lineage conversions: unnatural but useful? Nat Biotechnol 29:892–907. https://doi.org/10.1038/nbt.1946
Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M (2011) Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9:374–382. https://doi.org/10.1016/j.stem.2011.09.002
Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ, Furth EE, Sepulveda AR, Yuan CX, Won KJ, Donahue G, Sands J, Gumbs AA, Zaret KS (2013) An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep 3:2088–2099. https://doi.org/10.1016/j.celrep.2013.05.036
Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA (2008) In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455:627–632. https://doi.org/10.1038/nature07314
Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y (2012) Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11:100–109. https://doi.org/10.1016/j.stem.2012.05.018
Yu KR, Shin JH, Kim JJ, Koog MG, Lee JY, Choi SW, Kim HS, Seo Y, Lee S, Shin TH, Jee MK, Kim DW, Jung SJ, Shin S, Han DW, Kang KS (2015) Rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b. Cell Rep. https://doi.org/10.1016/j.celrep.2014.12.038
Matsui T, Takano M, Yoshida K, Ono S, Fujisaki C, Matsuzaki Y, Toyama Y, Nakamura M, Okano H, Akamatsu W (2012) Neural stem cells directly differentiated from partially reprogrammed fibroblasts rapidly acquire gliogenic competency. Stem Cells 30:1109–1119. https://doi.org/10.1002/stem.1091
Chung YG, Matoba S, Liu Y, Eum JH, Lu F, Jiang W, Lee JE, Sepilian V, Cha KY, Lee DR, Zhang Y (2015) Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 17:758–766. https://doi.org/10.1016/j.stem.2015.10.001
Cohnheim J (1875) Congenitales, Quergestreiftes Muskelsarkom der Nieren. Virchows Arch 65:64–69
Nguyen LV, Vanner R, Dirks P, Eaves CJ (2012) Cancer stem cells: an evolving concept. Nat Rev Cancer 12:133–143. https://doi.org/10.1038/nrc3184
Plaks V, Koopman CD, Werb Z (2013) Cancer. Circulating tumor cells. Science 341:1186–1188. https://doi.org/10.1126/science.1235226
Lin YC, Murayama Y, Hashimoto K, Nakamura Y, Lin CS, Yokoyama KK, Saito S (2014) Role of tumor suppressor genes in the cancer-associated reprogramming of human induced pluripotent stem cells. Stem Cell Res Ther 5:58. https://doi.org/10.1186/scrt447
Boulanger CA, Smith GH (2009) Reprogramming cell fates in the mammary microenvironment. Cell Cycle 8:1127–1132. https://doi.org/10.4161/cc.8.8.8189
Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K (2009) Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460:1145–1148. https://doi.org/10.1038/nature08285
Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, Brummelkamp TR (2010) Generation of iPSCs from cultured human malignant cells. Blood 115:4039–4042. https://doi.org/10.1182/blood-2009-07-231845
Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y, Mori M (2010) Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA 107:40–45. https://doi.org/10.1073/pnas.0912407107
Ramos-Mejia V, Fraga MF, Menendez P (2012) iPSCs from cancer cells: challenges and opportunities. Trends Mol Med 18:245–247. https://doi.org/10.1016/j.molmed.2012.04.001
Van Keymeulen A, Lee MY, Ousset M, Brohee S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I, Sotiriou C, Phillips WA, Blanpain C (2015) Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525:119–123. https://doi.org/10.1038/nature14665
Koren S, Reavie L, Couto JP, De Silva D, Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina O, Cardiff RD, Bentires-Alj M (2015) PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 525:114–118. https://doi.org/10.1038/nature14669
Ruetz T, Pfisterer U, Di Stefano B, Ashmore J, Beniazza M, Tian TV, Kaemena DF, Tosti L, Tan W, Manning JR, Chantzoura E, Ottosson DR, Collombet S, Johnsson A, Cohen E, Yusa K, Linnarsson S, Graf T, Parmar M, Kaji K (2017) Constitutively active SMAD2/3 are broad-scope potentiators of transcription-factor-mediated cellular reprogramming. Cell Stem Cell 21(791–805):e799. https://doi.org/10.1016/j.stem.2017.10.013
Kuo KK, Lee KT, Chen KK, Yang YH, Lin YC, Tsai MH, Wuputra K, Lee YL, Ku CC, Miyoshi H, Nakamura Y, Saito S, Wu CC, Chai CY, Eckner R, Steve Lin CL, Wang SS, Wu DC, Lin CS, Yokoyama KK (2016) Positive feedback loop of OCT4 and c-JUN expedites cancer stemness in liver cancer. Stem Cells 34:2613–2624. https://doi.org/10.1002/stem.2447
Wang SS, Wuputra K, Liu CJ, Lin YC, Chen YT, Chai CY, Lin CS, Kuo KK, Tsai MH, Wang SW, Chen KK, Miyoshi H, Nakamura Y, Saito S, Hanafusa T, Wu DC, Lin CS, Yokoyama KK (2016) Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget 7:36049–36064. https://doi.org/10.18632/oncotarget.9102
Fidler IJ, Kripke ML (1977) Metastasis results from preexisting variant cells within a malignant tumor. Science 197:893–895
Heppner GH (1984) Tumor heterogeneity. Cancer Res 44:2259–2265
Nowell PC (1986) Mechanisms of tumor progression. Cancer Res 46:2203–2207
Nowell PC (1976) The clonal evolution of tumor cell populations. Science 194:23–28
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11:726–734. https://doi.org/10.1038/nrc3130
Polyak K, Haviv I, Campbell IG (2009) Co-evolution of tumor cells and their microenvironment. Trends Genet 25:30–38. https://doi.org/10.1016/j.tig.2008.10.012
Morrissy AS, Cavalli FMG, Remke M, Ramaswamy V, Shih DJH, Holgado BL, Farooq H, Donovan LK, Garzia L, Agnihotri S, Kiehna EN, Mercier E, Mayoh C, Papillon-Cavanagh S, Nikbakht H, Gayden T, Torchia J, Picard D, Merino DM, Vladoiu M, Luu B, Wu X, Daniels C, Horswell S, Thompson YY, Hovestadt V, Northcott PA, Jones DTW, Peacock J, Wang X, Mack SC, Reimand J, Albrecht S, Fontebasso AM, Thiessen N, Li Y, Schein JE, Lee D, Carlsen R, Mayo M, Tse K, Tam A, Dhalla N, Ally A, Chuah E, Cheng Y, Plettner P, Li HI, Corbett RD, Wong T, Long W, Loukides J, Buczkowicz P, Hawkins CE, Tabori U, Rood BR, Myseros JS, Packer RJ, Korshunov A, Lichter P, Kool M, Pfister SM, Schuller U, Dirks P, Huang A, Bouffet E, Rutka JT, Bader GD, Swanton C, Ma Y, Moore RA, Mungall AJ, Majewski J, Jones SJM, Das S, Malkin D, Jabado N, Marra MA, Taylor MD (2017) Spatial heterogeneity in medulloblastoma. Nat Genet 49:780–788. https://doi.org/10.1038/ng.3838
Lu LC, Hsu CH, Hsu C, Cheng AL (2016) Tumor heterogeneity in hepatocellular carcinoma: facing the challenges. Liver Cancer 5:128–138. https://doi.org/10.1159/000367754
Shue YT, Lim JS, Sage J (2018) Tumor heterogeneity in small cell lung cancer defined and investigated in pre-clinical mouse models. Transl Lung Cancer Res 7:21–31. https://doi.org/10.21037/tlcr.2018.01.15
Cros J, Raffenne J, Couvelard A, Pote N (2018) Tumor heterogeneity in pancreatic adenocarcinoma. Pathobiology 85:64–71. https://doi.org/10.1159/000477773
Meacham CE, Morrison SJ (2013) Tumour heterogeneity and cancer cell plasticity. Nature 501:328–337. https://doi.org/10.1038/nature12624
Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I (2014) Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343:776–779. https://doi.org/10.1126/science.1247651
Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR (2014) Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509:371–375. https://doi.org/10.1038/nature13173
Halpern KB, Shenhav R, Matcovitch-Natan O, Toth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, Brandis A, Giladi A, Avihail AS, David E, Amit I, Itzkovitz S (2017) Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542:352–356. https://doi.org/10.1038/nature21065
Wang YJ, Schug J, Won KJ, Liu C, Naji A, Avrahami D, Golson ML, Kaestner KH (2016) Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65:3028–3038. https://doi.org/10.2337/db16-0405
Muraro MJ, Dharmadhikari G, Grun D, Groen N, Dielen T, Jansen E, van Gurp L, Engelse MA, Carlotti F, de Koning EJ, van Oudenaarden A (2016) A single-cell transcriptome atlas of the human pancreas. Cell Syst 3(385–394):e383. https://doi.org/10.1016/j.cels.2016.09.002
Barriga FM, Montagni E, Mana M, Mendez-Lago M, Hernando-Momblona X, Sevillano M, Guillaumet-Adkins A, Rodriguez-Esteban G, Buczacki SJA, Gut M, Heyn H, Winton DJ, Yilmaz OH, Attolini CS, Gut I, Batlle E (2017) Mex3a marks a slowly dividing subpopulation of Lgr5 + intestinal stem cells. Cell Stem Cell 20(801–816):e807. https://doi.org/10.1016/j.stem.2017.02.007
Johnson E, Dickerson KL, Connolly ID, Hayden Gephart M (2018) Single-cell RNA-sequencing in glioma. Curr Oncol Rep 20:42. https://doi.org/10.1007/s11912-018-0673-2
Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, Lee M, Hill D, Madden J, Boerner J, Cote ML, Sartor MA, Rozek LS, Wicha MS (2018) Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem Cell Rep 10:1596–1609. https://doi.org/10.1016/j.stemcr.2018.03.001
Giustacchini A, Thongjuea S, Barkas N, Woll PS, Povinelli BJ, Booth CAG, Sopp P, Norfo R, Rodriguez-Meira A, Ashley N, Jamieson L, Vyas P, Anderson K, Segerstolpe A, Qian H, Olsson-Stromberg U, Mustjoki S, Sandberg R, Jacobsen SEW, Mead AJ (2017) Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med 23:692–702. https://doi.org/10.1038/nm.4336
Prado K, Zhang KX, Pellegrini M, Chin AI (2017) Sequencing of cancer cell subpopulations identifies micrometastases in a bladder cancer patient. Oncotarget 8:45619–45625. https://doi.org/10.18632/oncotarget.17312
Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, Wong M, Choi PJ, Wee LJK, Hillmer AM, Tan IB, Robson P, Prabhakar S (2017) Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 49:708–718. https://doi.org/10.1038/ng.3818
Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ (2017) Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546:370–375. https://doi.org/10.1038/nature22403
Voog J, Jones DL (2010) Stem cells and the niche: a dynamic duo. Cell Stem Cell 6:103–115. https://doi.org/10.1016/j.stem.2010.01.011
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21:309–322. https://doi.org/10.1016/j.ccr.2012.02.022
Fessler E, Dijkgraaf FE, De Sousa EMF, Medema JP (2013) Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett 341:97–104. https://doi.org/10.1016/j.canlet.2012.10.015
Kim GA, Ginga NJ, Takayama S (2018) Integration of sensors in gastrointestinal organoid culture for biological analysis. Cell Mol Gastroenterol Hepatol 6(123–131):e121. https://doi.org/10.1016/j.jcmgh.2018.03.002
Dutta D, Heo I, Clevers H (2017) Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med 23:393–410. https://doi.org/10.1016/j.molmed.2017.02.007
Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, Nagae G, Ueda K, Nakazaki K, Kamikubo Y, Eto K, Aburatani H, Nakauchi H, Kurokawa M (2012) Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood 119:6234–6242. https://doi.org/10.1182/blood-2011-07-367441
Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H (2011) HIF induces human embryonic stem cell markers in cancer cells. Cancer Res 71:4640–4652. https://doi.org/10.1158/0008-5472.CAN-10-3320
Nagai K, Ishii H, Miyoshi N, Hoshino H, Saito T, Sato T, Tomimaru Y, Kobayashi S, Nagano H, Sekimoto M, Doki Y, Mori M (2010) Long-term culture following ES-like gene-induced reprogramming elicits an aggressive phenotype in mutated cholangiocellular carcinoma cells. Biochem Biophys Res Commun 395:258–263. https://doi.org/10.1016/j.bbrc.2010.03.176
Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R, Yuan Y, Gingold J, Xia W, Darr H, Mirzayans R, Hung MC, Schaniel C, Lemischka IR (2015) Modeling familial cancer with induced pluripotent stem cells. Cell 161:240–254. https://doi.org/10.1016/j.cell.2015.02.045
Zhou R, Xu A, Gingold J, Strong LC, Zhao R, Lee DF (2017) Li–Fraumeni syndrome disease model: a platform to develop precision cancer therapy targeting oncogenic p53. Trends Pharmacol Sci 38:908–927. https://doi.org/10.1016/j.tips.2017.07.004
Melo JV, Barnes DJ (2007) Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer 7:441–453. https://doi.org/10.1038/nrc2147
Chun YS, Byun K, Lee B (2011) Induced pluripotent stem cells and personalized medicine: current progress and future perspectives. Anat Cell Biol 44:245–255. https://doi.org/10.5115/acb.2011.44.4.245
Nair M, Sandhu SS, Sharma AK (2017) Induced pluripotent stem cell technology: a paradigm shift in medical science for drug screening and disease modeling. Curr Med Chem 24:4368–4398. https://doi.org/10.2174/0929867324666170727100508
Oshima N, Yamada Y, Nagayama S, Kawada K, Hasegawa S, Okabe H, Sakai Y, Aoi T (2014) Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS One 9:e101735. https://doi.org/10.1371/journal.pone.0101735
Nishi M, Akutsu H, Kudoh A, Kimura H, Yamamoto N, Umezawa A, Lee SW, Ryo A (2014) Induced cancer stem-like cells as a model for biological screening and discovery of agents targeting phenotypic traits of cancer stem cell. Oncotarget 5:8665–8680. https://doi.org/10.18632/oncotarget.2356
Nishi M, Sakai Y, Akutsu H, Nagashima Y, Quinn G, Masui S, Kimura H, Perrem K, Umezawa A, Yamamoto N, Lee SW, Ryo A (2014) Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene 33:643–652. https://doi.org/10.1038/onc.2012.614
Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, Liu JO, Deng C, Ye Z, Jang YY (2013) Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 57:2458–2468. https://doi.org/10.1002/hep.26237
Shyh-Chang N, Zheng Y, Locasale JW, Cantley LC (2011) Human pluripotent stem cells decouple respiration from energy production. EMBO J 30:4851–4852. https://doi.org/10.1038/emboj.2011.436
Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA (2012) Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11:589–595. https://doi.org/10.1016/j.stem.2012.10.005
Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A, Thangaraj K, Buchsbaum DJ, LoBuglio AF, Singh KK (2011) Cellular model of Warburg effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS One 6:e24792. https://doi.org/10.1371/journal.pone.0024792
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40:499–507. https://doi.org/10.1038/ng.127
Horimoto M, Resnick MB, Konkin TA, Routhier J, Wands JR, Baffy G (2004) Expression of uncoupling protein-2 in human colon cancer. Clin Cancer Res 10:6203–6207. https://doi.org/10.1158/1078-0432.CCR-04-0419
Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, Berggren WT, Zhang K, Evans RM, Siuzdak G, Izpisua Belmonte JC (2012) The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res 22:168–177. https://doi.org/10.1038/cr.2011.177
Lu J, Li H, Baccei A, Sasaki T, Gilbert DM, Lerou PH (2016) Influence of ATM-mediated DNA damage response on genomic variation in human induced pluripotent stem cells. Stem Cells Dev 25:740–747. https://doi.org/10.1089/scd.2015.0393
Corbet C (2017) Stem cell metabolism in cancer and healthy tissues: pyruvate in the limelight. Front Pharmacol 8:958. https://doi.org/10.3389/fphar.2017.00958
Sradhanjali S, Reddy MM (2018) Inhibition of pyruvate dehydrogenase kinase as a therapeutic strategy against cancer. Curr Top Med Chem 18:444–453. https://doi.org/10.2174/1568026618666180523105756
Pulito C, Donzelli S, Muti P, Puzzo L, Strano S, Blandino G (2014) microRNAs and cancer metabolism reprogramming: the paradigm of metformin. Ann Transl Med 2:58. https://doi.org/10.3978/j.issn.2305-5839.2014.06.03
Cantoria MJ, Boros LG, Meuillet EJ (2014) Contextual inhibition of fatty acid synthesis by metformin involves glucose-derived acetyl-CoA and cholesterol in pancreatic tumor cells. Metabolomics 10:91–104. https://doi.org/10.1007/s11306-013-0555-4
Larue L, Bellacosa A (2005) Epithelial–mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene 24:7443–7454. https://doi.org/10.1038/sj.onc.1209091
Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, Maestro R, Voeltzel T, Selmi A, Valsesia-Wittmann S, Caron de Fromentel C, Puisieux A (2008) Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14:79–89. https://doi.org/10.1016/j.ccr.2008.06.005
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial–mesenchymal transitions in development and disease. Cell 139:871–890. https://doi.org/10.1016/j.cell.2009.11.007
Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Pollan M, Linares J, Serrano S, Saez-Castillo AI, Sanchez L, Pajares R, Sanchez-Aguilera A, Artiga MJ, Piris MA, Rodriguez-Peralto JL (2007) A high-throughput study in melanoma identifies epithelial–mesenchymal transition as a major determinant of metastasis. Cancer Res 67:3450–3460. https://doi.org/10.1158/0008-5472.CAN-06-3481
Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Sudhof TC (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798. https://doi.org/10.1016/j.neuron.2013.05.029
Nebbioso A, Tambaro FP, Dell’Aversana C, Altucci L (2018) Cancer epigenetics: moving forward. PLoS Genet 14:e1007362. https://doi.org/10.1371/journal.pgen.1007362
Villanueva MT (2015) Epigenetics: chromatin marks the spot. Nat Rev Cancer 15:196–197. https://doi.org/10.1038/nrc3934
Sun L, Fang J (2016) Epigenetic regulation of epithelial–mesenchymal transition. Cell Mol Life Sci 73:4493–4515. https://doi.org/10.1007/s00018-016-2303-1
Trager MM, Dhayat SA (2017) Epigenetics of epithelial-to-mesenchymal transition in pancreatic carcinoma. Int J Cancer 141:24–32. https://doi.org/10.1002/ijc.30626
Toh TB, Lim JJ, Chow EK (2017) Epigenetics in cancer stem cells. Mol Cancer 16:29. https://doi.org/10.1186/s12943-017-0596-9
Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, Chiou SH, Schep AN, Baral J, Hamard C, Antoine M, Wislez M, Kong CS, Connolly AJ, Park KS, Sage J, Greenleaf WJ, Winslow MM (2016) Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166:328–342. https://doi.org/10.1016/j.cell.2016.05.052
Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA (2010) Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467:1114–1117. https://doi.org/10.1038/nature09515
De S, Ganesan S (2017) Looking beyond drivers and passengers in cancer genome sequencing data. Ann Oncol 28:938–945. https://doi.org/10.1093/annonc/mdw677
Pon JR, Marra MA (2015) Driver and passenger mutations in cancer. Annu Rev Pathol 10:25–50. https://doi.org/10.1146/annurev-pathol-012414-040312
Gonzalez-Perez A, Mustonen V, Reva B, Ritchie GR, Creixell P, Karchin R, Vazquez M, Fink JL, Kassahn KS, Pearson JV, Bader GD, Boutros PC, Muthuswamy L, Ouellette BF, Reimand J, Linding R, Shibata T, Valencia A, Butler A, Dronov S, Flicek P, Shannon NB, Carter H, Ding L, Sander C, Stuart JM, Stein LD, Lopez-Bigas N, International Cancer Genome Consortium Mutation P, Consequences Subgroup of the Bioinformatics Analyses Working G (2013) Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods 10:723–729. https://doi.org/10.1038/nmeth.2562
Meyerson M, Gabriel S, Getz G (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11:685–696. https://doi.org/10.1038/nrg2841
Watson IR, Takahashi K, Futreal PA, Chin L (2013) Emerging patterns of somatic mutations in cancer. Nat Rev Genet 14:703–718. https://doi.org/10.1038/nrg3539
Kato S, Lippman SM, Flaherty KT, Kurzrock R (2016) The conundrum of genetic “Drivers” in benign conditions. J Natl Cancer Inst. https://doi.org/10.1093/jnci/djw036
Korkaya H, Liu S, Wicha MS (2011) Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Investig 121:3804–3809. https://doi.org/10.1172/JCI57099
Wang B, Miyagoe-Suzuki Y, Yada E, Ito N, Nishiyama T, Nakamura M, Ono Y, Motohashi N, Segawa M, Masuda S, Takeda S (2011) Reprogramming efficiency and quality of induced Pluripotent Stem Cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr 3:RRN1274. https://doi.org/10.1371/currents.RRN1274
Broxmeyer HE (2010) Will iPS cells enhance therapeutic applicability of cord blood cells and banking? Cell Stem Cell 6:21–24. https://doi.org/10.1016/j.stem.2009.12.008
Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7:64–77. https://doi.org/10.1016/j.stem.2010.04.015
Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7:51–63. https://doi.org/10.1016/j.stem.2010.04.014
Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385:810–813. https://doi.org/10.1038/385810a0
Buganim Y, Faddah DA, Jaenisch R (2013) Mechanisms and models of somatic cell reprogramming. Nat Rev Genet 14:427–439. https://doi.org/10.1038/nrg3473
Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J (2013) NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495:370–374. https://doi.org/10.1038/nature11925
Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA (1999) The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res 27:2291–2298
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, Guan KL, Xiong Y (2013) Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32:663–669. https://doi.org/10.1038/onc.2012.67
Di Croce L, Helin K (2013) Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20:1147–1155. https://doi.org/10.1038/nsmb.2669
Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, Martinez-Garcia E, Zhang H, Zheng Y, Verma SK, McCabe MT, Ott HM, Van Aller GS, Kruger RG, Liu Y, McHugh CF, Scott DW, Chung YR, Kelleher N, Shaknovich R, Creasy CL, Gascoyne RD, Wong KK, Cerchietti L, Levine RL, Abdel-Wahab O, Licht JD, Elemento O, Melnick AM (2013) EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23:677–692. https://doi.org/10.1016/j.ccr.2013.04.011
Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22:5323–5335. https://doi.org/10.1093/emboj/cdg542
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100:11606–11611. https://doi.org/10.1073/pnas.1933744100
Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP (2008) Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40:741–750. https://doi.org/10.1038/ng.159
Funato K, Major T, Lewis PW, Allis CD, Tabar V (2014) Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346:1529–1533. https://doi.org/10.1126/science.1253799
Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340:857–861. https://doi.org/10.1126/science.1232245
Khan SN, Jankowska AM, Mahfouz R, Dunbar AJ, Sugimoto Y, Hosono N, Hu Z, Cheriyath V, Vatolin S, Przychodzen B, Reu FJ, Saunthararajah Y, O’Keefe C, Sekeres MA, List AF, Moliterno AR, McDevitt MA, Maciejewski JP, Makishima H (2013) Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia 27:1301–1309. https://doi.org/10.1038/leu.2013.80
Simon JA, Kingston RE (2013) Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49:808–824. https://doi.org/10.1016/j.molcel.2013.02.013
Smith LL, Yeung J, Zeisig BB, Popov N, Huijbers I, Barnes J, Wilson AJ, Taskesen E, Delwel R, Gil J, Van Lohuizen M, So CW (2011) Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell 8:649–662. https://doi.org/10.1016/j.stem.2011.05.004
Yuan J, Takeuchi M, Negishi M, Oguro H, Ichikawa H, Iwama A (2011) Bmi1 is essential for leukemic reprogramming of myeloid progenitor cells. Leukemia 25:1335–1343. https://doi.org/10.1038/leu.2011.85
Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G (2009) BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci 29:8884–8896. https://doi.org/10.1523/JNEUROSCI.0968-09.2009
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. https://doi.org/10.1146/annurev.biochem.77.062706.153223
Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, Cassel SH, Qing H, Widmer CJ, Demetri GD, Irizarry RA, Zhao K, Ranish JA, Kadoch C (2017) SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat Genet 49:1613–1623. https://doi.org/10.1038/ng.3958
Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, Tzvetkov EP, Troisi EC, Pomeroy SL, Biegel JA, Tolstorukov MY, Bernstein BE, Park PJ, Roberts CW (2017) SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 49:289–295. https://doi.org/10.1038/ng.3746
Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, Park PJ, Shivdasani RA, Roberts CW (2017) ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet 49:296–302. https://doi.org/10.1038/ng.3744
Pombo A, Dillon N (2015) Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16:245–257. https://doi.org/10.1038/nrm3965
Achinger-Kawecka J, Taberlay PC, Clark SJ (2016) Alterations in three-dimensional organization of the cancer genome and epigenome. Cold Spring Harb Symp Quant Biol 81:41–51. https://doi.org/10.1101/sqb.2016.81.031013
Long HK, Prescott SL, Wysocka J (2016) Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167:1170–1187. https://doi.org/10.1016/j.cell.2016.09.018
Herz HM (2016) Enhancer deregulation in cancer and other diseases. BioEssays 38:1003–1015. https://doi.org/10.1002/bies.201600106
Risca VI, Greenleaf WJ (2015) Unraveling the 3D genome: genomics tools for multiscale exploration. Trends Genet 31:357–372. https://doi.org/10.1016/j.tig.2015.03.010
Shlyueva D, Stampfel G, Stark A (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15:272–286. https://doi.org/10.1038/nrg3682
Wang C, Huang S (2014) Nuclear function of Alus. Nucleus 5:131–137. https://doi.org/10.4161/nucl.28005
Jin C, Felsenfeld G (2007) Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21:1519–1529. https://doi.org/10.1101/gad.1547707
Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41:941–945. https://doi.org/10.1038/ng.409
Fagnocchi L, Poli V, Zippo A (2018) Enhancer reprogramming in tumor progression: a new route towards cancer cell plasticity. Cell Mol Life Sci 75:2537–2555. https://doi.org/10.1007/s00018-018-2820-1
Pulecio J, Verma N, Mejia-Ramirez E, Huangfu D, Raya A (2017) CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell 21:431–447. https://doi.org/10.1016/j.stem.2017.09.006
Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, Li Y, Vershkov D, Cacace A, Young RA, Jaenisch R (2018) Rescue of Fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172(979–992):e976. https://doi.org/10.1016/j.cell.2018.01.012
Zezulin A, Musunuru K (2018) Turning up the heat with therapeutic epigenome editing. Cell Stem Cell 22:10–11. https://doi.org/10.1016/j.stem.2017.12.013
Pflueger C, Tan D, Swain T, Nguyen T, Pflueger J, Nefzger C, Polo JM, Ford E, Lister R (2018) A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. https://doi.org/10.1101/gr.233049.117
Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Zhao Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H (2008) Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 3:475–479. https://doi.org/10.1016/j.stem.2008.10.002
Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460:1149–1153. https://doi.org/10.1038/nature08287
Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460:1136–1139. https://doi.org/10.1038/nature08290
Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460:1140–1144. https://doi.org/10.1038/nature08311
Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R (2009) Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462:595–601. https://doi.org/10.1038/nature08592
Liao J, Marumoto T, Yamaguchi S, Okano S, Takeda N, Sakamoto C, Kawano H, Nii T, Miyamato S, Nagai Y, Okada M, Inoue H, Kawahara K, Suzuki A, Miura Y, Tani K (2013) Inhibition of PTEN tumor suppressor promotes the generation of induced pluripotent stem cells. Mol Ther 21:1242–1250. https://doi.org/10.1038/mt.2013.60
Kwon D, Kim JS, Cha BH, Park KS, Han I, Park KS, Bae H, Han MK, Kim KS, Lee SH (2016) The effect of fetal bovine serum (FBS) on efficacy of cellular reprogramming for induced pluripotent stem cell (iPSC) generation. Cell Transplant 25:1025–1042. https://doi.org/10.3727/096368915X689703
Sarig R, Rivlin N, Brosh R, Bornstein C, Kamer I, Ezra O, Molchadsky A, Goldfinger N, Brenner O, Rotter V (2010) Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J Exp Med 207:2127–2140. https://doi.org/10.1084/jem.20100797
Chen CL, Wang LJ, Yan YT, Hsu HW, Su HL, Chang FP, Hsieh PC, Hwang SM, Shen CN (2014) Cyclin D1 acts as a barrier to pluripotent reprogramming by promoting neural progenitor fate commitment. FEBS Lett 588:4008–4017. https://doi.org/10.1016/j.febslet.2014.08.039
Jiang H, Xu Z, Zhong P, Ren Y, Liang G, Schilling HA, Hu Z, Zhang Y, Wang X, Chen S, Yan Z, Feng J (2015) Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat Commun 6:10100. https://doi.org/10.1038/ncomms10100
Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN (2009) Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 15:501–513. https://doi.org/10.1016/j.ccr.2009.03.018
Liu L, Wise DR, Diehl JA, Simon MC (2008) Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem 283:31153–31162. https://doi.org/10.1074/jbc.M805056200
Van Blerkom J (2009) Mitochondria in early mammalian development. Semin Cell Dev Biol 20:354–364. https://doi.org/10.1016/j.semcdb.2008.12.005
Varum S, Momcilovic O, Castro C, Ben-Yehudah A, Ramalho-Santos J, Navara CS (2009) Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res 3:142–156. https://doi.org/10.1016/j.scr.2009.07.002
Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan W, Wang M, Yang W, Pei G (2014) Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res 24:665–679. https://doi.org/10.1038/cr.2014.32
Keith B, Simon MC (2007) Hypoxia-inducible factors, stem cells, and cancer. Cell 129:465–472. https://doi.org/10.1016/j.cell.2007.04.019
Saito S, Lin YC, Tsai MH, Lin CS, Murayama Y, Sato R, Yokoyama KK (2015) Emerging roles of hypoxia-inducible factors and reactive oxygen species in cancer and pluripotent stem cells. Kaohsiung J Med Sci 31:279–286. https://doi.org/10.1016/j.kjms.2015.03.002
Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, Chen K, Li Y, Liu X, Xu J, Zhang S, Li F, He W, Labuda K, Song Y, Peterbauer A, Wolbank S, Redl H, Zhong M, Cai D, Zeng L, Pei D (2010) Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6:71–79. https://doi.org/10.1016/j.stem.2009.12.001
Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M (1997) Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol 17:3094–3102
Tanigawa S, Lee CH, Lin CS, Ku CC, Hasegawa H, Qin S, Kawahara A, Korenori Y, Miyamori K, Noguchi M, Lee LH, Lin YC, Steve Lin CL, Nakamura Y, Jin C, Yamaguchi N, Eckner R, Hou DX, Yokoyama KK (2013) Jun dimerization protein 2 is a critical component of the Nrf2/MafK complex regulating the response to ROS homeostasis. Cell Death Dis 4:e921. https://doi.org/10.1038/cddis.2013.448
Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT (1998) Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA 95:11715–11720
Plaks V, Kong N, Werb Z (2015) The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16:225–238. https://doi.org/10.1016/j.stem.2015.02.015
Pavlides S, Tsirigos A, Vera I, Flomenberg N, Frank PG, Casimiro MC, Wang C, Pestell RG, Martinez-Outschoorn UE, Howell A, Sotgia F, Lisanti MP (2010) Transcriptional evidence for the “Reverse Warburg Effect” in human breast cancer tumor stroma and metastasis: similarities with oxidative stress, inflammation, Alzheimer’s disease, and “Neuron-Glia Metabolic Coupling”. Aging (Albany NY) 2:185–199. https://doi.org/10.18632/aging.100134
Wong GS, Rustgi AK (2013) Matricellular proteins: priming the tumour microenvironment for cancer development and metastasis. Br J Cancer 108:755–761. https://doi.org/10.1038/bjc.2012.592
Ye J, Wu D, Wu P, Chen Z, Huang J (2014) The cancer stem cell niche: cross talk between cancer stem cells and their microenvironment. Tumour Biol 35:3945–3951. https://doi.org/10.1007/s13277-013-1561-x
Smith KN, Starmer J, Miller SC, Sethupathy P, Magnuson T (2017) Long noncoding RNA moderates MicroRNA activity to maintain self-renewal in embryonic stem cells. Stem Cell Rep 9:108–121. https://doi.org/10.1016/j.stemcr.2017.05.005
Kaufhold S, Garban H, Bonavida B (2016) Yin Yang 1 is associated with cancer stem cell transcription factors (SOX2, OCT4, BMI1) and clinical implication. J Exp Clin Cancer Res 35:84. https://doi.org/10.1186/s13046-016-0359-2
Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Gray NS, Young RA (2017) YY1 is a structural regulator of enhancer-promoter loops. Cell 171(1573–1588):e1528. https://doi.org/10.1016/j.cell.2017.11.008
Ifkovits JL, Addis RC, Epstein JA, Gearhart JD (2014) Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One 9:e89678. https://doi.org/10.1371/journal.pone.0089678
Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin C, Fu JD, Wang GG, Liu J, Qian L (2016) Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 18:382–395. https://doi.org/10.1016/j.stem.2016.02.003
Zhou H, Morales MG, Hashimoto H, Dickson ME, Song K, Ye W, Kim MS, Niederstrasser H, Wang Z, Chen B, Posner BA, Bassel-Duby R, Olson EN (2017) ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev 31:1770–1783. https://doi.org/10.1101/gad.305482.117
Julian LM, McDonald AC, Stanford WL (2017) Direct reprogramming with SOX factors: masters of cell fate. Curr Opin Genet Dev 46:24–36. https://doi.org/10.1016/j.gde.2017.06.005
Wapinski OL, Lee QY, Chen AC, Li R, Corces MR, Ang CE, Treutlein B, Xiang C, Baubet V, Suchy FP, Sankar V, Sim S, Quake SR, Dahmane N, Wernig M, Chang HY (2017) Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep 20:3236–3247. https://doi.org/10.1016/j.celrep.2017.09.011
Yaqubi M, Mohammadnia A, Fallahi H (2015) Predicting involvement of polycomb repressive complex 2 in direct conversion of mouse fibroblasts into induced neural stem cells. Stem Cell Res Ther 6:42. https://doi.org/10.1186/s13287-015-0045-x
Zhang QJ, Li JJ, Lin X, Lu YQ, Guo XX, Dong EL, Zhao M, He J, Wang N, Chen WJ (2017) Modeling the phenotype of spinal muscular atrophy by the direct conversion of human fibroblasts to motor neurons. Oncotarget 8:10945–10953. https://doi.org/10.18632/oncotarget.14641
Kochat V, Equbal Z, Baligar P, Kumar V, Srivastava M, Mukhopadhyay A (2017) JMJD3 aids in reprogramming of bone marrow progenitor cells to hepatic phenotype through epigenetic activation of hepatic transcription factors. PLoS One 12:e0173977. https://doi.org/10.1371/journal.pone.0173977
Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, Cowin BL, Bruner E, Murphy MM, Chen W, Spangrude GJ, Hu Z, DeBerardinis RJ, Morrison SJ (2017) Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549:476–481. https://doi.org/10.1038/nature23876
Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, Grillo I, Bakogianni S, Ndiaye-Lobry D, Martin MT, Guillamot M, Banh RS, Xu M, Figueroa ME, Dickins RA, Abdel-Wahab O, Park CY, Tsirigos A, Neel BG, Aifantis I (2017) Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170(1079–1095):e1020. https://doi.org/10.1016/j.cell.2017.07.032
Cimmino L, Neel BG, Aifantis I (2018) Vitamin C in stem cell reprogramming and cancer. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2018.04.001
Gustafson CB, Yang C, Dickson KM, Shao H, Van Booven D, Harbour JW, Liu ZJ, Wang G (2015) Epigenetic reprogramming of melanoma cells by vitamin C treatment. Clin Epigenetics 7:51. https://doi.org/10.1186/s13148-015-0087-z
Bershteyn M, Kriegstein AR (2013) Cerebral organoids in a dish: progress and prospects. Cell 155:19–20. https://doi.org/10.1016/j.cell.2013.09.010
Dutta D, Clevers H (2017) Organoid culture systems to study host-pathogen interactions. Curr Opin Immunol 48:15–22. https://doi.org/10.1016/j.coi.2017.07.012
Cornu TI, Mussolino C, Cathomen T (2017) Refining strategies to translate genome editing to the clinic. Nat Med 23:415–423. https://doi.org/10.1038/nm.4313
Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J (2018) CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. https://doi.org/10.1038/s41591-018-0049-z
Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, Kaykas A (2018) p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 24:939–946. https://doi.org/10.1038/s41591-018-0050-6
Weeber F, Ooft SN, Dijkstra KK, Voest EE (2017) Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol 24:1092–1100. https://doi.org/10.1016/j.chembiol.2017.06.012
Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T (2006) Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun 340:183–189. https://doi.org/10.1016/j.bbrc.2005.11.164
Wang LJ, Zhang H, Wang YS, Xu WB, Xiong XR, Li YY, Su JM, Hua S, Zhang Y (2011) Scriptaid improves in vitro development and nuclear reprogramming of somatic cell nuclear transfer bovine embryos. Cell Reprogram 13:431–439. https://doi.org/10.1089/cell.2011.0024
Fan N, Chen J, Shang Z, Dou H, Ji G, Zou Q, Wu L, He L, Wang F, Liu K, Liu N, Han J, Zhou Q, Pan D, Yang D, Zhao B, Ouyang Z, Liu Z, Zhao Y, Lin L, Zhong C, Wang Q, Wang S, Xu Y, Luan J, Liang Y, Yang Z, Li J, Lu C, Vajta G, Li Z, Ouyang H, Wang H, Wang Y, Yang Y, Liu Z, Wei H, Luan Z, Esteban MA, Deng H, Yang H, Pei D, Li N, Pei G, Liu L, Du Y, Xiao L, Lai L (2013) Piglets cloned from induced pluripotent stem cells. Cell Res 23:162–166. https://doi.org/10.1038/cr.2012.176
Pandey A, Gupta SC, Singh N, Rana JS, Gupta N (2010) Efficiency of SCNT buffalo (Bubalus bubalis) embryos in different culture medium and analysis of mRNA expression of insulin-like growth factors during embryogenesis. Reprod Domest Anim 45:786–795. https://doi.org/10.1111/j.1439-0531.2009.01353.x
Huang Y, Tang X, Xie W, Zhou Y, Li D, Zhou Y, Zhu J, Yuan T, Lai L, Pang D, Ouyang H (2011) Vitamin C enhances in vitro and in vivo development of porcine somatic cell nuclear transfer embryos. Biochem Biophys Res Commun 411:397–401. https://doi.org/10.1016/j.bbrc.2011.06.160
Mallol A, Santalo J, Ibanez E (2015) Improved development of somatic cell cloned mouse embryos by vitamin C and latrunculin A. PLoS One 10:e0120033. https://doi.org/10.1371/journal.pone.0120033
Xiong X, Li J, Wang L, Zhong J, Zi X, Wang Y (2014) Low oxygen tension and relative defined culture medium with 3,4-dihydroxyflavone are beneficial for yak-bovine interspecies somatic cell nuclear transfer embryo. Reprod Domest Anim 49:126–133. https://doi.org/10.1111/rda.12240
Kumar BM, Maeng GH, Lee YM, Lee JH, Jeon BG, Ock SA, Kang T, Rho GJ (2013) Epigenetic modification of fetal fibroblasts improves developmental competency and gene expression in porcine cloned embryos. Vet Res Commun 37:19–28. https://doi.org/10.1007/s11259-012-9542-x
Huan YJ, Zhu J, Xie BT, Wang JY, Liu SC, Zhou Y, Kong QR, He HB, Liu ZH (2013) Treating cloned embryos, but not donor cells, with 5-aza-2′-deoxycytidine enhances the developmental competence of porcine cloned embryos. J Reprod Dev 59:442–449
Sun L, Wu KL, Zhang D, Wang HY, Wang Y, Xu ZY, Huang XY, Chen ZJ, Liu HQ (2012) Increased cleavage rate of human nuclear transfer embryos after 5-aza-2′-deoxycytidine treatment. Reprod Biomed Online 25:425–433. https://doi.org/10.1016/j.rbmo.2012.06.018
Kunitomi A, Yuasa S, Sugiyama F, Saito Y, Seki T, Kusumoto D, Kashimura S, Takei M, Tohyama S, Hashimoto H, Egashira T, Tanimoto Y, Mizuno S, Tanaka S, Okuno H, Yamazawa K, Watanabe H, Oda M, Kaneda R, Matsuzaki Y, Nagai T, Okano H, Yagami KI, Tanaka M, Fukuda K (2016) H1foo has a pivotal role in qualifying induced pluripotent stem cells. Stem Cell Rep 6:825–833. https://doi.org/10.1016/j.stemcr.2016.04.015
Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454:49–55. https://doi.org/10.1038/nature07056
Lee J, Xia Y, Son MY, Jin G, Seol B, Kim MJ, Son MJ, Do M, Lee M, Kim D, Lee K, Cho YS (2012) A novel small molecule facilitates the reprogramming of human somatic cells into a pluripotent state and supports the maintenance of an undifferentiated state of human pluripotent stem cells. Angew Chem Int Ed Engl 51:12509–12513. https://doi.org/10.1002/anie.201206691
Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA (2008) Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26:1269–1275. https://doi.org/10.1038/nbt.1502
Maherali N, Hochedlinger K (2009) Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol 19:1718–1723. https://doi.org/10.1016/j.cub.2009.08.025
Huntly BJ, Gilliland DG (2005) Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer 5:311–321. https://doi.org/10.1038/nrc1592
Chen M, Huang J, Yang X, Liu B, Zhang W, Huang L, Deng F, Ma J, Bai Y, Lu R, Huang B, Gao Q, Zhuo Y, Ge J (2012) Serum starvation induced cell cycle synchronization facilitates human somatic cells reprogramming. PLoS One 7:e28203. https://doi.org/10.1371/journal.pone.0028203
Deng W (2010) AID in reprogramming: quick and efficient: identification of a key enzyme called AID, and its activity in DNA demethylation, may help to overcome a pivotal epigenetic barrier in reprogramming somatic cells toward pluripotency. BioEssays 32:385–387. https://doi.org/10.1002/bies.201000014
Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM (2010) Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463:1042–1047. https://doi.org/10.1038/nature08752
Popowski M, Templeton TD, Lee BK, Rhee C, Li H, Miner C, Dekker JD, Orlanski S, Bergman Y, Iyer VR, Webb CF, Tucker H (2014) Bright/Arid3A acts as a barrier to somatic cell reprogramming through direct regulation of Oct4, Sox2, and Nanog. Stem Cell Rep 2:26–35. https://doi.org/10.1016/j.stemcr.2013.12.002
Chen T, Yuan D, Wei B, Jiang J, Kang J, Ling K, Gu Y, Li J, Xiao L, Pei G (2010) E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells 28:1315–1325. https://doi.org/10.1002/stem.456
Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S (2009) Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 5:237–241. https://doi.org/10.1016/j.stem.2009.08.001
Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S (2013) Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater 12:1154–1162. https://doi.org/10.1038/nmat3777
Gruber R, Panayiotou R, Nye E, Spencer-Dene B, Stamp G, Behrens A (2016) YAP1 and TAZ control pancreatic cancer initiation in mice by direct up-regulation of JAK-STAT3 signaling. Gastroenterology 151:526–539. https://doi.org/10.1053/j.gastro.2016.05.006
Ahmad F, Patrick S, Sheikh T, Sharma V, Pathak P, Malgulwar PB, Kumar A, Joshi SD, Sarkar C, Sen E (2017) Telomerase reverse transcriptase (TERT)—enhancer of zeste homolog 2 (EZH2) network regulates lipid metabolism and DNA damage responses in glioblastoma. J Neurochem 143:671–683. https://doi.org/10.1111/jnc.14152
Liu X, Huang Q, Li F, Li CY (2014) Enhancing the efficiency of direct reprogramming of human primary fibroblasts into dopaminergic neuron-like cells through p53 suppression. Sci China Life Sci 57:867–875. https://doi.org/10.1007/s11427-014-4730-2
Jin F, Wang Y, Zhu Y, Li S, Liu Y, Chen C, Wang X, Zen K, Li L (2017) The miR-125a/HK2 axis regulates cancer cell energy metabolism reprogramming in hepatocellular carcinoma. Sci Rep 7:3089. https://doi.org/10.1038/s41598-017-03407-3
Outani H, Okada M, Yamashita A, Nakagawa K, Yoshikawa H, Tsumaki N (2013) Direct induction of chondrogenic cells from human dermal fibroblast culture by defined factors. PLoS One 8:e77365. https://doi.org/10.1371/journal.pone.0077365
Zhao Z, Xu M, Wu M, Ma K, Sun M, Tian X, Zhang C, Fu X (2015) Direct reprogramming of human fibroblasts into sweat gland-like cells. Cell Cycle 14:3498–3505. https://doi.org/10.1080/15384101.2015.1093707
Kwon D, Ji M, Lee S, Seo KW, Kang KS (2017) Reprogramming enhancers in somatic cell nuclear transfer, iPSC technology, and direct conversion. Stem Cell Rev 13:24–34. https://doi.org/10.1007/s12015-016-9697-x
Acknowledgements
We thank W.H. Lin for editing this manuscript and H. Niwa for helpful comments. This work was supported partially by grants from the Ministry of Science and Technology (MOST 106-2320-B-037-001-MY3, MOST 106-2320-B-037-028, and MOST 106-2314-B-037-017), by the National Health Research Institutes (NHRI-Ex107-10720SI); and Kaohsiung Medical University grants (KMU-TP103A04, KMU-TP103G03, KMU-TP104E23, and KMU-DT104001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Saito, S., Lin, YC., Nakamura, Y. et al. Potential application of cell reprogramming techniques for cancer research. Cell. Mol. Life Sci. 76, 45–65 (2019). https://doi.org/10.1007/s00018-018-2924-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2924-7