Abstract
Biological organisms can pose a threat to water quality and can be responsible for waterborne diseases. Because it is very expensive and time consuming to test for each pathogenic organism, coliform bacteria and E. coli, which originate in environmental and animal sources, serve as good pathogenic indicator organisms. This study focused on evaluation of water quality in New Jersey water bodies by monitoring both E. coli and coliform using different methods. Water samples were taken over two months at different sites. Three different methods used in this study are membrane filtration, PetrifilmTM and IDEXX methods to compare and determine which one or in combination methods are the most efficient and reliable way to identify and quantify bacterial contamination. It was concluded that areas that were highly urbanized, specifically high urban and agriculturally developed areas, had an increased level of contamination. Our study suggested that modified membrane filter method in combination of IDEXX Colilert® System provide the best results. Further research, such as Microbial Source Tracking (MST), will be conducted to identify the origin of fecal contamination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
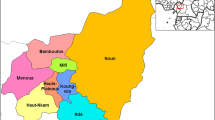
Northern New Jersey has diverse array of land use and land cover patterns. Much of Northern New Jersey is encompassed within the Passaic River Basin, which is one of the most flood-prone watersheds in the United States [1]. The watershed is 935 square miles, and encompasses 9 counties in Northern New Jersey and Southern New York State. The basin is divided into three main regions based on topography: the Highlands, the Central Basin, and the Lower Valley. The collection sites chosen for this study all fall within this watershed. The Highlands region comprises the most upstream portions of the Passaic River. Its land use is composed of mainly agriculture and forested lands [2]. This portion of the Passaic River is designated for primary contact use, which allows for activities such as water sports and activities where ingestion and immersion are likely [3]. The Central Basin region is characterized by 262 square miles of low-lying flat land composed of meadows, bogs, and swamps [1]. Like the Highlands region, this portion of the river is designated for primary contact [3]. The Lower Valley encompasses the most downstream portion of the Passaic River from Little Falls to where it empties into Newark Bay. This is the most developed region of the river system; it includes urban areas such as Paterson, Passaic, and Newark [1]. The Lower Valley region of the Passaic River includes a highly polluted area called “Chemical Row,” [4]. The Lower Passaic River also contains over 70 combined sewer overflows, which allow wastewater to enter the river, along with excess rainwater, during storm events [5].
Although water bodies used for drinking water, recreational activities, or food production are monitored for many types of pollutions, one of particular concern is fecal contamination. Fecal contamination of rivers and lakes is common around the world, regardless of the level of urbanization or economic status of the region. All countries experience this pollution, although the type and degree vary [6].
Fecal contamination is a great concern because it may harbor both bacterial and viral pathogens able to threaten human health, such as hepatitis A virus, Vibrio species, Salmonella species, Campylobacter, Norwalk group viruses [7], poliovirus, coxsackievirus [5], Escherichia coli O157:H17, Mycobacterium species, and Listeria species [5], [8]. An essential step in preventing outbreaks such as those mentioned above in the future is the detection, quantification, and identification of fecal contaminants.
Traditional methods of measuring the presence of indicator bacteria involve the total coliform membrane-filter technique endorsed by the Environmental Protection Agency [9] and the American Public Health Association. This method involves the use of the selective medium modified Endo agar (m-Endo agar), which allows for growth of only coliform bacteria.
A different method for detection and quantification of E. coli and total coliforms is provided by IDEXX’s Colilert® system (IDEXX laboratories). This system is recommended by the Environmental Protection Agency for testing ambientwaters [10]. This method is selective, like the total coliform membrane filter technique. It uses IDEXX’s Defined Substrate Technology®, where nutrient indicators metabolize into differently colored Products by E. coli or total coliforms. Another method that involves growing and counting coliform colonies uses 3M Petrifilm™ E. coli/Coliform Count Plates (3M, United States). This method is approved by the United States Food and Drug Administration for testing all foods and milk. However, at this time it is not approved but only recommended for water testing purposes. Once bacterial species have been cultured from environmental sample, via any of the methods mentioned above, further culturing techniques were used to grow and maintain the bacteria. The bacterial were then being used to quantitatively verifying the above methods. The micro source tracking was also attempted in one site to detect possible source of contamination.
II. Methods
A. Sample Collection and Processing
Seven study sites were selected within the Passaic River Watershed (Table 1 and Fig. 1). River samples were collected on March 9, 2013 and April 13, 2013. Samples collection methodologies were based upon the recommendations from USEPA [11]. At each designated site, samples were collected in heat-resistant plastic 1 liter bottles, which had been previously autoclaved. During April samplings, a 100 ml sterile cup sample was also collected. At the time of collections air and water temperatures were recorded. All samples were kept on ice until they were returned to the laboratory for processing.
Once in the laboratory, samples were processed the same day, within 6 hours of collections. Samples first underwent vacuum filtration through grade 1 Whatman 12.5 cm, 11 μm pore filters to remove large debris, dirt, etc. The filtrate then underwent vacuum filtration through a Thermo Scientific Nalgene® Cellulose Nitrate (CN) Filter Unit; 150 ml, 0.45 μm Pore Size from Cole-Parmer, or Nalgene MF75 Vacuum Filter Units PES Membrane 1,000 ml 0.2 μm 90 mm, Complete Units with Supor or machV Membrane (Voigt Global Distribution). The filters were cut into four parts with sterile scissors. Three of the one fourth of each filter was placed into three microfuge tubes with sterile tweezers. The tubes were filled with 1 ml water and vortexed for about 2 minutes. One sample was used for M-Endo agar plate analysis. One sample was used for 3M Petrifilm™ E. coli/ Coliform Count Plate. The plates and Petrifilm were incubated at 37°C for 24 hours. The M-Endo plates were observed for metallic sheen and the number of colonies. E. coli and coliform colonies were selected from 3M Petrifilm™ based upon their coloration. Blue colonies with gas indicated E. coli and red and blue colonies with gas indicated total coliform. One fourth of the filter was suspended as described and put into 99 ml of sterilized deionized water for IDEXX Colilert® analysis [12](IDEXX Laboratories, Inc., ME, USA). Portions of the filter that were not used the same day were stored in sterile foil in the freezer for further molecular analysis. The colonies collected from M-Endo and 3M Petrifilm™ were inoculated aseptically into nutrient broth or on nutrient agar plate. The plates or broth were incubated for 24 hours at 37°C. The cultures were used as quantitative control to verify the results from different methods.
B. Chelex Method of DNA Extraction and PCR-based Assay
The Chelex DNA extraction was used for LP2 sample. One quarter of the filter was cut with sterile scissors and placed into a microfuge tube with sterile tweezers, along with 200 μl 5% Chelex solution. The tubes were vortexed for 30 seconds. They were incubated in a 56°C water bath for 2 hours, and then vortexed again for 10 seconds. The tubes were then placed into a boiling water bath for 8 minutes, and then vortexed for 10 seconds. The tubes were centrifuged for 3 minutes at 10,000 rpm and the supernatants were placed into new tubes, NanoDrop ND-1000 spectrophotometer which was used to determine the DNA quality and concentration (ng/μl) of DNA extraction product [13].
General primers for total coliforms, and E. coli [14], [15], [16]; specific primer for human [17] and Canadian geese [18] were selected for PCR based assay to identify indicator organisms and source of the fecal pollution respectively. The target organisms, expected amplicon size, and melting temperature (°C) of each primer are shown in Table 2. The reaction mix with primers, DNA and Hot-Start Taq Mastermix were prepared and run in an Applied Biosystems Veriti 96 Well Thermocycler (Life Technologies). All PCR products were visualized through 1% agarose gel electrophoresis with a Hi-Lo DNA Ladder as a marker.
III. Results and Discussion
A. Three Identification and Quantitation Methods
Three quantitative methods were used to analyze the environmental samples collected. These methods were Modified Total Coliform Membrane Filter Technique, IDEXX’s Colilert® System, and Modified 3M™ Petrifilm™. The air and water temperatures for March and April were showed in table 3. From the total coliform membrane filter techniques (M-Endo) analysis, the March samples had limited or non-detectable E. coli and Coliform. The April samples showed the Lower Valley region had 51.2 in site LP1, 4.8 in site GF1, and 1.6 in site LP2 respectively. The Highlands region showed 14.4 total coliform CFU per 100 ml at site GT1, 1.6 at site GT2, 0.4 at site UP2, and although site UP1 had metallic coloration, no colonies were visible. There are larger numbers in the lower valley region (sites LP1, GF1, LP2) than in the highlands region (sites UP1, GT1, UP2) except in GT2 (Fig. 2).
IDEXX’s Colilert® System was able to identify the presence of E. coli in all of the April samples (Fig. 3). The Highlands samples yielded 1.6 (site GT2) to 54.4 (site GT1) E. coli per 100 ml of sample. The number of total coliforms in this group ranged from 53.6 (site GT2) to 150.3 (site GT1) per 100 ml of sample. E. coli and total coliforms were proportionate; site GT2 had the least for both categories, followed by site UP2, and site GT1 had the most for both categories. The Lower Valley samples also showed in that the sites that had the most total coliforms also had the most E. coli. Site GF1 had the highest numbers, with 131.7 E. coli and 1,046.2 total coliforms per 100 ml. Site LP2 followed with 91.4 E. coli and 223.5 total coliform, and site LP1 had the lowest numbers for the Lower Valley, with 82.2 E. coli and 156.4 total coliforms.
3M Petrifilm™ methods indicated that the collected water samples all had greater numbers of total coliforms in April than in March. The highlands sites, UP1, GT1, GT2, and UP2, all had zero positive results in March, but had numbers ranging from 3.2 (site UP1) to 27.2 (site GT1) in April. The lower valley sites ranged from 23.6 (site GF1) to 30.4 (site LP1) in April (Fig. 4). Thus, the lower valley region tended to have higher counts of total coliforms, and the numbers tended to be higher in April.
It was expected that data derived from these three methods would correlate, since they are all quantitative methods, but our data demonstrated inconsistency among three methods. However, the general trend of more bacteria being present in the lower valley region as opposed to the highlands region was concluded from this study. In addition, it was shown that in April, water contained more E. coli and Coliform than March collection. The order of sites from most to least contamination was different according to the different methods. The sites were ordered GF1, LP2 and LP1 according to IDEXX; LP1, LP2, GF1 according to 3M™ Petrifilm™ data, and LP1, GF1, LP2 according to M-Endo plate data.
In order to evaluate the quantitative accuracy of these methodologies, serial dilutions (10-3 to 10-6) of cultures obtained from purified colonies of coliforms and E. coli were used to test each methodology. The Modified Total Coliform Membrane Filter technique yielded the best results. The m- Endo plates allowed accurately observe the dilutions visualized as colonies on the plates. The serial dilutions did not show correlated counts from IDEXX data due to the limitation of its level of detection (the maximum level of detection is 2,419 per 100 ml) (IDEXX Laboratories, Inc., ME, USA). The 3M Petrifilm™ inoculated with all four dilutions developed a dense layer of bubbles that affect the counting of the bacterial. This indicates that the maximum detectable CFU is less than m-Endo method.
Previous studies indicated that IDEXX’s Colilert® system is as accurate as or more accurate than traditional methodologies [19], [20], and it is a USEPA recommended method (IDEXX Laboratories, Inc., ME, USA). Thus, our recommendation for future studies is that IDEXX’s Colilert system be combined with total coliform membrane filter technique, to ensure that the samples are within the range of the system (1-2,149 CFU per 100 ml).
B. Pathogen Indicators Analysis
Our results indicated that all sites tested positive for both E. coli and total coliforms in April samplings. According to drinking water standards, the presence of E. coli in these samples would over the maximum contaminant level (MCL) [21]. Thus, this water would not be suitable for drinking water, and is currently not used for this purpose.
Primary and secondary recreational contact is designated according to E. coli levels. All April river samples tested positive for E. coli, with the numbers ranging from 1 to 131 CFU per 100 ml. In New Jersey, primary contact is currently limited to water bodies that have averagely less than 126 CFU per 100 ml and no single detection contain greater than 235 CFU per 100 ml. Secondary contact is limited to waters with less than 770 CFU of fecal coliform per 100 ml. According to our data, sites GT1, GT2, UP2, LP1, and LP2 all fall within the acceptable range for primary contact set by the EPA (United States Environmental Protection Agency, 2012). These sites all have values of less than 126 CFU E. coli per 100 ml. Currently, designations agree with our classification of sites UP1, UP2, LP1, and LP2, as primary contact is authorized in all parts of the Passaic River upstream of the Dundee Dam. Our data indicated that site GF1 might be unsuitable for primary contact. At 131 CFU E. coli per 100 ml, this value does not exceed the maximum concentration allowed for a single sample, less than 235 CFU per 100 ml, but if this concentration is typical, the geometric mean (GM) of this site may be above the maximum allowed, 126 CFU E. coli per 100 ml. More samplings would need to be performed in order to determine the GM of this site.
Additionally, samples tested in this study were taken in spring. Due to the seasonal variation, it is anticipated colony forming pathogen populations will greatly increase during the host summer months. Therefore, additional testing in summer is required to determine the suitability of water to be used for human recreational purposes.
C. PCR Identification
The sample from the site of LP2 was used as an example of molecular identification. Specific primers of E. coli, coliform and Micro source tracking of geese and human were used for PCR based assay (data not shown). PCR products from the primer set 27F/1492R were expected to have a band about 1,460 bp in length if E. coli were present. PCR products from the primer set LZL389/LZL653 were expected to have a band around 264 bp in length if samples contained coliforms. Samples showed positive results for both E. coli and Coliform. Some specific sets of primers were used for micro source tracking to identify the source of contamination. The results suggested that the possibility of human fecal pollution in sample LP2 and a positive band with the expected amplicon size of 77 bp suggested Canadian Geese fecal contamination. Further studies should be conducted to confirm these results.
IV. Conclusion
Membrane filter techniques, 3M Petrifilm™ and IDEXX’s Colilert® System were used in this study. Our evaluation indicates that IDEXX’s Colilert® System should be used in conjunction with modified Total Coliform Membrane Filter Technique in order to confirm that levels are within the detectable range of the Colilert® System. After the data iscompiled, all methods showed a similar trend with regard to E. coli and Coliform population. It seems that the April samples had higher counts of coliforms and the Lower Valley region seemed to have more fecal contamination than the Highlands region. Preliminary data using gene-specific PCR-based assay suggested source of contamination due to human or geese pollution, since the area is densely populated and contains combined sewer overflows (CSOs).
Tables and Figures
References
The Passaic River Coalition, “What We Do - Education - Passaic River Basin,” 2010; http://www.passaicriver.org/passaicriverinfo.html.
K.C. Ponader, D.F. Charles, and T.J. Belton, “Diatom-based TP and TN inference models and indices for monitoring nutrient enrichment of New Jersey streams,” Ecological Indicators, vol. 7, no. 1, 2007, pp. 79–93.
New Jersey Department of Environmental Protection (NJDEP), “New Jersey Surface Water Quality Standards,” 2008, pp. 1–113.
S.L. Huntley, R.J. Wenning, S.H. Su, N.L. Bonnevie, and D.J. Paustenbach, “Geochronology and sedimentology of the lower Passaic River, New Jersey,” Estuaries, vol. 18, no. 2, 1995, pp. 351–361.
E. Donovan, K. Unice, J.D. Roberts, M. Harris, and B. Finley, “Risk of gastrointestinal disease associated with exposure to pathogens in the water of the Lower Passaic River,” Appl Environ Microbiol, vol. 74, no. 4, 2008, pp. 994–1003.
P. Roslev, and A.S. Bukh, “State of the art molecular markers for fecal pollution source tracking in water,” Appl Microbiol Biotechnol, vol. 89, no. 5, 2011, pp. 1341–1355.
H.J. Ma, L.L. Fu, and J.R. Li, “Differentiation of fecal Escherichia coli from human, livestock, and poultry sources by rep-PCR DNA fingerprinting on the shellfish culture area of East China Sea,” Curr Microbiol, vol. 62, no. 5, 2011, pp. 1423–1430.
K. Murugan, P. Prabhakaran, S. Al-Sohaibani, and K. Sekar, “Identification of source of faecal pollution of Tirumanimuttar River, Tamilnadu, India using microbial source tracking,” Environ Monit Assess, vol. 184, no. 10, 2011, pp. 6001–6012.
A.E. Greenberg, “Standard Methods: For the Examination of Water and Wastewater, 18th Edition,” 18th ed., A.E. Greenberg, L.S. Clescerl, and A.D. Eaton eds., Amer Public Health Assn, 1992, pp. 9222A: 9229-9253; 9222B: 9229-9254; 9222C: 9229–9258.
E. O’Brien, “Volunteers conduct bacteria methods comparison study,” 2006, pp. 1–24.
United States Environmental Protection Agency (USEPA), “Method 1604: Total Coliforms and Escherichia coli in Water by Membrane Filtration Using a Simultaneous Detection Technique (MI Medium) ” 2002, pp. 18.
D. Miller, “Quanti-Tray and Quanti-Tray/2000 IDEXX MPN Generator 3.2.,” 2005
T.C. Chu, and M.J. Rienzo, “Bloom-Forming Cyanobacteria and Other Phytoplankton in Northern New Jersey Freshwater Bodies,” International Perspectives on Water Quality Management and Pollutant Control, W.T. Nigel ed., InTech Publishing, 2013, pp. 1–23.
A.K. Bej, M.H. Mahbubani, J.L. Dicesare, and R.M. Atlas, “Polymerase chain reaction-gene probe detection of microorganisms by using filter-concentrated samples,” Appl Environ Microbiol, vol. 57, no. 12, 1991, pp. 3529–3534.
D.J. Lane, “16S/23S rRNA sequencing,” Nucleic Acid Techniques in Bacterial Systematics, E. Stackerbrandt, and M. Goodfellow eds., John Wiley & Sons Ltd., 1991, pp. 115–147.
S. Turner, K.M. Pryer, V.P. Miao, and J.D. Palmer, “Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis,” J Eukaryot Microbiol, vol. 46, no. 4, 1999, pp. 327–338.
A.E. Bernhard, and K.G. Field, “A PCR assay To discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA,” Appl Environ Microbiol, vol. 66, no. 10, 2000, pp. 4571–4574.
J.M. Caldwell, and J.F. Levine, “Domestic wastewater influent profiling using mitochondrial real-time PCR for source tracking animal contamination,” J Microbiol Methods, vol. 77, no. 1, 2009, pp. 17–22.
K.F. Eckner, “Comparison of membrane filtration and multiple-tube fermentation by the colilert and enterolert methods for detection of waterborne coliform bacteria, Escherichia coli, and enterococci used in drinking and bathing water quality monitoring in southern sweden,” Appl Environ Microbiol, vol. 64, no. 8, 1998, pp. 3079–3083.
G.P. Yakub, D.A. Castric, K.L. Stadterman-Knauer, M.J. Tobin, M. Blazina, T.N. Heineman, G.Y. Yee, and L. Frazier, “Evaluation of Colilert and Enterolert defined substrate methodology for wastewater applications,” Water Environ Res, vol. 74, no. 2, 2002, pp. 131–135.
United States Environmental Protection Agency (USEPA), “2012 Recreational Water Quality Criteria,” EPA-820-F-12-061, 2012
Author information
Authors and Affiliations
Corresponding author
Additional information
Lee H. Lee, Meiyin Wu, Alexandra Peri and Tin-Chun Chu
Manuscript received December 31, 2013. This work was supported by Montclair State University Faculty Scholarship Program (FSP) to LHL and MW; Seton Hall University Biological Sciences Research Fund to TC.
L. H. Lee is a Professor at the Department of Biology & Molecular Biology, Montclair State University, Montclair, NJ 07043 USA (e-mail: Lee.Lee@montclair.edu).
M. Wu is the Director of the Passaic River Institute and Professor of the Department of Biology & Molecular Biology, Montclair State University, Montclair, NJ 07043 USA (e-mail: wum@mail.montclair.edu).
A. Peri was a graduate student at the Department of Biology & Molecular Biology, Montclair State University, Montclair, NJ 07043 USA (e-mail: PeriA1@mail.montclair.edu).
T. C. Chu is an Associate Professor of the Department of Biological Sciences, Seton Hall University, South Orange, NJ 07079 USA (e-mail: Tin-Chun.Chu@shu.edu).
Dr. Lee H. Lee received her B.S. in Plant Pathology and Entomology from National Taiwan University, and her M.S. and Ph.D. in Biology from the City University of New York. She is a professor in the department of Biology and Molecular Biology at Montclair State University. Dr. Lee’s research interest is in Environmental Microbiology. She has carried out studies of microorganisms that can act as environmental pollution indicators for water bodies in New Jersey. She has published more than 45 journal articles and member for many professional societies including American Society for Microbiology.
Dr. Meiyin Wu, Director of Passaic River Institute and Professor of Biology and Molecular Biology at Montclair State University, is an ecologist whose research focuses on habitat restoration and ecosystem management. Dr. Wu’s research emphasizes on species invasion, aquatic ecology and management, and ecological restoration. Dr. Wu uses a variety of field research and laboratory analytical techniques to examine the interactions in ecosystems. Additionally, she provides environmental education opportunities and stakeholder outreach programs to a very diverse audience groups. She is the author of more than 30 articles, books and DVDs and holds two U.S. patents with another patent pending.
Dr. Tin-Chun Chu received her B.S. in Molecular Biology from Montclair State University and her Ph.D. in Biomedical Informatics from University of Medicine and Dentistry of New Jersey. She is a molecular microbiologist with bioinformatics expertise. She is currently an Associate Professor of Biological Sciences at Seton Hall University. One of her research interests is on the ecological dynamics of the harmful bloom-forming cyanobacteria and their phages. Since joining SHU in fall 2008, she has published more than 20 peer-reviewed journal articles and book chapters. She holds one U.S. patents with another pending.
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, L., Wu, M., Peri, A. et al. Method Evaluations for Escherichia coli and Coliforms Detection in Northern New Jersey Water Bodies. GSTF J Biosci 3, 2 (2014). https://doi.org/10.7603/s40835-014-0002-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.7603/s40835-014-0002-y