Abstract
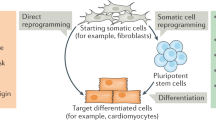
Direct epigenetic reprogramming is a technique that converts a differentiated adult cell into another differentiated cell—such fibroblasts to cardiomyocytes—without passage through an undifferentiated pluripotent stage. This novel technology is opening doors in biological research and regenerative medicine. Some preliminary studies about direct reprogramming started in the 1980s when differentiated adult cells could be converted into other differentiated cells by overexpressing transcription-factor genes. These studies also showed that differentiated cells have plasticity. Direct reprogramming can be a powerful tool in biological research and regenerative medicine, especially the new frontier of personalized medicine. This review aims to summarize all direct reprogramming studies of somatic cells by master control genes as well as potential applications of these techniques in research and treatment of selected human diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cell fate and reprogramming
The human body originates from a totipotent stem cell, the zygote. Development and growth of an organism are due to proliferation and differentiation of these cells. Stem cell proliferation by self-renewal causes an increase in cell numbers, while stem cell differentiation causes an increase in cell types. Although all cells in the human body originate from a single cell, they play different roles. Their finalized specific functions are decided by mechanisms that are yet unclear, but it is considered that their functions are decided by their fates or programming (alterations in gene expression). From a single totipotent stem cell, generations of daughter cells are programmed into specific cell types that collaborate with each other to produce a completed body.
In the traditional view, cell fates cannot be modified, and stem cell differentiation is unidirectional, in which only uncommitted or undifferentiated cells can differentiate into committed or specific cells. However, to date, many studies prove that fully differentiated cells can reverse to pluripotent stem cells. This process is termed as “reprogramming” (Fig. 1 ).
Reprogramming
The first attempt of the reprogramming technique was performed by Robert Briggs and Thomas King. In 1952, they injected an embryonic nucleus into an enucleated egg in the amphibian Rana pipiens (Briggs and King, 1952), advancing from an oocyte to the tadpole stage of development. However, this experiment failed when carried out with fully differentiated cells. These results made them conclude that differentiated nuclei cannot revert to a developing embryo (King and Briggs, 1955). Conversely, John B. Gurdon successfully produced swimming tadpoles from transplantation of differentiated tadpole intestinal epithelial cell nuclei into enucleated eggs that were exposed to ultraviolet irradiation (Gurdon, 1962). By this result, Gurdon concluded that differentiated somatic cell nuclei can revert to pluripotency.
Gurdon’s discovery opened a new field in animal development biology. He presented a new mechanism that changed the opinion of hundreds of biological scientists. This discovery was confirmed by Wilmut et al. in 1997. Similarly, for the first time in a mammal, Wilmut successfully created the sheep Dolly by injecting adult mammary epithelial cell nuclei into an enucleated sheep egg (Wilmut et al., 1997). After these results, more than 10 different species, such as mouse, cow, pig, cat, and dog, have been “cloned” by the injection adult cell nuclei into oocytes, and the technique is also called “somatic cell nuclear transfer—SCNT.”
Although Gurdon showed that differentiated cell nuclei could be reprogrammed into the undifferentiated state to re-start development, many experiments also suggested that the intact, differentiated cells could be reprogrammed into undifferentiated cells. This was confirmed by Shinya Yamanaka in 2007. He choose 24 transcription factors related to embryonic stem cells, and from these 24 genes his group demonstrated that only four genes, Myc, Oct3/4, Sox2, and Klf4 could reprogram mouse embryonic fibroblasts into pluripotent stem cells (Takahashi and Yamanaka, 2006). These stem cells were termed as induced pluripotent stem cells (iPSCs) by Yamanaka. iPSCs exhibit most of the characteristics of embryonic stem cells such as selfrenewal and long-term-multiple lineage differentiation, and have been especially useful in the production of mouse chimeras. In 2007, Yamanaka and James Thomson’s laboratories were the first to successfully produce human iPSCs (Takahashi et al., 2007a; Yu et al., 2007). Yamanaka’s group used the four factors found in mouse: Oct4, Sox2, Klf4, and Myc (OSKM); Thomson used the set: Lin28, Nanog, Oct4, and Sox2. For these critical contributions in reprogramming technology, John Gurdon and Shinya Yamanaka shared a Nobel Prize in Physiology and Medicine in 2012.
Since 2006, iPSC technology has been continuously refined to produce iPSCs with higher efficiency and easier and safer production. In a study, OSKM was transfected in mouse embryonic fibroblasts by viral vectors (Takahashi et al., 2007b). Four of these factors would activate the pluripotent status of differentiated cells (Jaenisch and Young, 2008). In human fibroblasts, Oct4 and Sox2, together with Nanog and LIN28, can reprogram them toward pluripotent cells (Yu et al., 2007).
To improve the efficiency of reprogramming, subsequent studies used polycistronic vector containing four factors, chromatin-modifying chemicals, and mRNAs, in combination with activation or inhibition of various signaling pathways involved in the regulation of cell proliferation (Chang et al., 2009; Feng et al., 2009; Heinrich and Dimmeler, 2012; Kretsovali et al., 2012).
Some studies also significantly improved the safety of transgenes. In an early study, retroviral vectors were integrated into a genome, causing insertional mutagenesis. This technique is can be modified by utilizing non-integrating vectors (Stadtfeld and Hochedlinger, 2010; Stadtfeld et al., 2008). Further advances related to DNA-free transgenes using mRNAs or proteins were achieved (Jia et al., 2010; Warren et al., 2010; Zhou et al., 2009).
With these improvements, clinical-grade iPSCs were developed in the recent years. Clinical grade iPSCs usually use donor cells such as fibroblasts, keratinocytes, and peripheral blood mononuclear cells (PBMCs), which are preferable for inducing pluripotency. Moreover, clinical-grade iPSCs need to be produced from safer techniques, reducing the likelihood of accidently creating tumor-forming cells.
Some safer techniques in gene transfection are used to produce vectors containing reprogramming genes. The first effort used F-deficient Sendai virus particles to induce pluripotency in somatic cells (Dowey et al., 2012; Fusaki et al., 2009). iPSCs produced using this method must be sub-cultured for 10–20 passages to remove the excess virus particles and to make virusfree iPSC lines. Later, an improvement in gene transfection using temperature-sensitive Sendai virus particles made it is easier to remove the virus particles by temperature shift (Ban et al., 2011).
Virus-free vectors carrying reprogramming factors have been studied since 2010 to replace the viral vectors. Episomal DNA can be used to transfect transgenes into adult cells. These virus-free vectors have important clinical applications because they are safer in manipulations as well as in the patients. There are two kinds of episomes: non-replicating episomal vectors and replicating episomal vectors. The iPSC production procedure using non-replicating episomal vectors is of low-yield; therefore, multiple transfections are suggested as a solution to increase the iPSC production efficacy (Jia et al., 2010; Okita et al., 2008). Improvements such as the use of minicircle or codonoptimized 4-in-1 minicircle (CoMiP) DNA vectors were devised (Lu et al., 2013; Okita et al., 2008).
Although DNA-based episome is considered safe to reprogram adult cells to iPSCs, in principle, foreign DNA can integrate into the host genome. Therefore, iPSCs must be screened to select free cells for further applications (Gonzalez et al., 2009). To date, the safest technique of iPSC production is induction of pluripotency via mRNA (Warren et al., 2012; Yoshioka et al., 2013) or protein (Kim et al., 2009; Lee et al., 2012). These iPSCs are called “clean” iPSCs.
Together with improvement of iPSC production methods, some approaches using iPSCs in treatment were also developed. The most significant approach for clinical applications of iPSCs relates to the combination of iPSC technology and targeting editing of the iPSC genome. This combination helps to push iPSCs into clinical treatment, particularly for patients with genetic disorders. There are three ways to correct the mutated genes in iPSCs: the zinc finger nuclease (ZFN) system, the transcription activator-like effector nuclease (TALEN) system, and the clustered regularly interspaced short palindromic repeats (CRISPR) system (Ding et al., 2013; Hockemeyer et al., 2009; Horii et al., 2013). By using these techniques, patient-specific iPSCs were successfully produced to treat epilepsy (Parent and Anderson, 2015), myotonic dystrophy type 1 (Xia et al., 2015), sickle erythrocytes (Huang et al., 2015), retinal degenerative diseases (Wiley et al., 2015), and recessive dystrophic epidermolysis bullosa (Sebastiano et al., 2014).
Direct reprogramming
The direct reprogramming technique was discovered in the 1980s (Table 1 ). In 1987, Davis et al. converted embryonic mouse fibroblasts into muscle cells by transfection of myogenic differentiation factor (MyoD) (Davis et al., 1987). Similarly, MyoD was used to reprogram immature chondrocytes, smooth muscle cells, and retinal cells into muscle cells (Choi et al., 1990). In the 1990s, some other transcription factors were discovered, particularly globin transcription factor 1 (Gata-1), that can reprogram avian monocyte precursors into erythrocytes, eosinophils, and megakaryocytes (Kulessa et al., 1995).
Since 2000, several transcription factors were discovered and were successfully used to reprogram target cells such as pancreatic islet cells (Zhou et al., 2008), neurons (Fishman et al., 2015; Vierbuchen et al., 2010), hepatocytes (Huang et al., 2011; Sekiya and Suzuki, 2011), endothelial cells (Ginsberg et al., 2012; Han et al., 2014), smooth muscle cells (Karamariti et al., 2013), and hepatocyte like cells (Simeonov and Uppal, 2014).
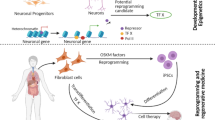
In recent years, in situ direct reprogramming as well as in vivo direct reprogramming has become important, as the ability to provide novel therapies is nearly in clinical applications. In vivo direct reprogramming is the usage of specific transcription factors to change target cell fate in the body without the need to isolate the target cells (Table 1 ).
In early studies, it was shown that transcription factors can directly affect reprogramming. Recent studies indicated that there are at least five kinds of reprogramming factors that can directly reprogram adult cells into other phenotypic cells: transcription factors, epigenetic regulators, miRNAs, Small molecules, and pluripotency factors for direct reprogramming.
Transcription factors
Different from reprogramming techniques make adult cells pluripotent after receiving some key transcription factors causing epigenetic modifications, direct reprogramming mechanisms are still elusive. The most important mechanism is the effect of transcription factors that drive the phenotype changes in specific cells. By using transcription factors, transfected cells can change phenotype via activation of target genes. Interestingly, these changes can occur some hours after transfections (Ieda et al., 2010), do not require cell division (Heinrich et al., 2010; Vierbuchen et al., 2010), and are stable after removal of reprogramming factors (Huang et al., 2011; Sekiya and Suzuki, 2011). Some authors have demonstrated that direct reprogramming of fibroblasts to neurons was hierarchical, established mechanisms dictate that fibroblasts gradually change with multiple steps to become neurons (Wapinski et al., 2013).
Epigenetic regulators
Differentiated status of cells seems depend on epigenetic status of these cells. Transcription factors are known as important factors effecting to expression of lineage specific genes. However, gene expression also is effected by epigenetic regulators. In fact, there are three ways that epigenetic regulators effect gene expression. First, epigenetic regulators can decide the reprogramming process by themselves. For example, pancreatic beta cells can be reprogrammed into alpha cells by DNA methyltransferase Dnmt1 deficiency (Dhawan et al., 2011). Second, epigenetic regulators can interact with exogenous factors to re-activate or suppress related gene expression. In the study by Takeuchi and Bruneau (2009), they showed that Baf60c – cardiac specific subunit of BAF chromatin remodeling complexes hold a particular role in the reprogramming from mouse mesoderm to cardiac myocytes that is helped by Gata4 – a transcription factor to bind to cardiac genes (Takeuchi and Bruneau, 2009). Third, some epigenetic regulators act as epigenetic barriers that can prevent reprogramming. In fact, the inhibition or removal of histone deacetylases and polycomb repressor complex 2 (PRC2) can facilitate the reprogramming of germ cells into neurons (Patel et al., 2012).
miRNAs
More and more studies proved that miRNAs play important roles in the reprogramming process. Some specific miRNAs such as miR-124, miR-9/9, miR-1, miR-133, miR-208, and miR-499 were demonstrated with reprogramming effects in fibroblasts. Overexpression of miR-9/9 and miR-124 in human fibroblasts can induce the expression of markers indicative of neuron-like cells (Yoo et al., 2011). It seems that miRNAs can regulate some mechanisms relating to epigenetic reprogramming. In fact, miRNAs can directly stimulate or suppress target genes (Bartel, 2009) as well as regulate epigenetic regulators (Neo et al., 2014). However, in general, miRNAs are not as efficient as transcription factors to induce epigenetic reprogramming.
Small molecules
Some small molecules were successfully used to produce iPSC (Li et al., 2013b). The main advantage of small molecules is small structure, therefore they can more easily move across cellular membranes. By this advantage, small molecules are more richly investigated in recent studies. The biggest success in direct reprogramming by small molecules is the neural conversion process (Kim et al., 2014; Sayed et al., 2015). How the small molecules can reprogram the cell fate is a question that needs to be answered. In some cases, small molecules activate some pluripotency genes (Hou et al., 2013) as well as transcription factors (Yuan et al., 2013).
Pluripotency Factors for Indirect Reprogramming
Some pluripotency factors used to produce iPSC can directly reprogram some cell types such as cardiomyocytes (Efe et al., 2011), neural stem cells or progenitors (Wang et al., 2013), angioblast-like progenitor cells (Kurian et al., 2013), endothelial cells (Li et al., 2013a), pancreatic lineages (Li et al., 2014), and hepatocytes (Zhu et al., 2014). Ma et al. (2013) showed that pluripotent factors can reprogram adult cells into pluripotent cells with multiple steps and that at certain steps some cells’ fates are formed as transition stages of epigenetic reprogramming (Ma et al., 2013). Moreover, overexpression of pluripotent factors can also induce differentiation (Loh and Lim, 2011).
Although direct reprogramming can produce the functional cells that can be used in translational applications as well as therapy, the main limitation of this technology is slow or non-proliferation of reprogrammed cells. Therefore, direct reprogramming should be improved in order to produce proliferating cells such as tissue specific stem cells or progenitor cells more than fully differentiated cells. In fact, some kinds of stem cells as well as progenitor cells were produced by direct reprogramming technology, including neural stem cells or progenitors (Han et al., 2012; Schindeler et al., 2015; Thier et al., 2012), oligodendrocyte precursor cells (Najm et al., 2013), hepatic stem cells (Yu et al., 2013), HSCs (Riddell et al., 2014), and hematopoietic multipotent progenitors (Batta et al., 2014; Sandler et al., 2014).
Invivo direct reproramming
As direct reprogramming technology is gradually perfected, especially its efficiency in combination with the tools of in situ gene therapy that were developed in previous studies. In vivo direct reprogramming has become more interesting as a novel therapy in regenerative medicine. Using in situ gene therapy strategies with direct reprogramming factors, some preclinical trials with a mouse model were successful in the conversion of various cerebral cell types into neurons (Heinrich and Rouaux, 2015). By enhanced expression of Sox10 in Satellite Glial cells, Weider et al (2015) successfully induced these cells in vivo into oligodendrocyte- like cells (Weider et al., 2015).
Particularly, reactive glial cells in the cortex of stabinjured or Alzheimer’s disease (AD) model mice can be directly reprogrammed into functional neurons in vivo using retroviral expression of a single neural transcription factor, NeuroD1 (Guo et al., 2014). More importantly, cardiac injury model mice can be treated by in vivo direct reprogramming(Jayawardena et al., 2015). miRNAs and lentiviral vectors were injected into these mice. After 5-6 weeks, cardiac function was improved, associated with existence of cardiac myocyte- like cells in injected sites.
Conclusion
Epigenetic reprogramming has seen rapid growth in recent years. Supported by some modern molecular biology techniques, reprogramming technology is becoming important and promising for wide use in basic research to translational research, and clinical application in the near future. Direct epigenetic reprogramming is a combination of stem cell therapy and gene therapy that can induce cell regeneration in an in situ manner. Many non-viral vectors and some novel reprogramming factors have facilitated direct reprogramming applications in preclinical models. Direct reprogramming, however, also faces with some challenges. Safety of vectors as well as technology must be investigated and carefully evaluated, especially in viral vector transfections or DNA transfection. Another challenge relates to control of reprogramming efficiency as well as specificity of target cells in vivo.
Abbreviations
AD: Alzheimer’s disease; PRC2: Polycomb repressor complex 2; ZFN: zinc finger nuclease; HSCs: Hematopoietic stem cells; iPSC: Induced pluripotent stem cells.
Acknowledgement
This work is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106-YS.06-2013.37.
Competing interests
The authors declare that they have no competing interests.
References
Adler, A.F., Grigsby, C.L., Kulangara, K., Wang, H., Yasuda, R., and Leong, K.W. (2012). Nonviral direct conversion of primary mouse embryonic fibroblasts to neuronal cells. Molecular therapy Nucleic acids 1, e32.
Ban, H., Nishishita, N., Fusaki, N., Tabata, T., Saeki, K., Shikamura, M., Takada, N., Inoue, M., Hasegawa, M., Kawamata, S., et al. (2011). Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proceedings of the National Academy of Sciences of the United States of America 108, 14234–14239.
Bartel, D.P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233.
Batta, K., Florkowska, M., Kouskoff, V., and Lacaud, G. (2014). Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell reports 9, 1871–1884.
Berninger, B., Costa, M.R., Koch, U., Schroeder, T., Sutor, B., Grothe, B., and Gotz, M. (2007). Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 8654–8664.
Bichsel, C., Neeld, D., Hamazaki, T., Chang, L.J., Yang, L.J., Terada, N., and Jin, S. (2013). Direct reprogramming of fibroblasts to myocytes via bacterial injection of MyoD protein. Cellular reprogramming 15, 117–125.
Briggs, R., and King, T.J. (1952). Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proceedings of the National Academy of Sciences of the United States of America 38, 455–463.
Bussmann, L.H., Schubert, A., Vu Manh, T.P., De Andres, L., Desbordes, S.C., Parra, M., Zimmermann, T., Rapino, F., Rodriguez- Ubreva, J., Ballestar, E., et al. (2009). A robust and highly efficient immune cell reprogramming system. Cell stem cell 5, 554–566.
Caiazzo, M., Dell’Anno, M.T., Dvoretskova, E., Lazarevic, D., Taverna, S., Leo, D., Sotnikova, T.D., Menegon, A., Roncaglia, P., Colciago, G., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227.
Chang, C.W., Lai, Y.S., Pawlik, K.M., Liu, K., Sun, C.W., Li, C., Schoeb, T.R., and Townes, T.M. (2009). Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem cells (Dayton, Ohio) 27, 1042–1049.
Choi, J., Costa, M.L., Mermelstein, C.S., Chagas, C., Holtzer, S., and Holtzer, H. (1990). MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proceedings of the National Academy of Sciences of the United States of America 87, 7988–7992.
Davis, R.L., Weintraub, H., and Lassar, A.B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000.
Dhawan, S., Georgia, S., Tschen, S.I., Fan, G., and Bhushan, A. (2011). Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Developmental cell 20, 419–429.
Ding, Q., Lee, Y.K., Schaefer, E.A., Peters, D.T., Veres, A., Kim, K., Kuperwasser, N., Motola, D.L., Meissner, T.B., Hendriks, W.T., et al. (2013). A TALEN genome-editing system for generating human stem cell-based disease models. Cell stem cell 12, 238–251.
Dowey, S.N., Huang, X., Chou, B.K., Ye, Z., and Cheng, L. (2012). Generation of integration-free human induced pluripotent stem cells from postnatal blood mononuclear cells by plasmid vector expression. Nature protocols 7, 2013–2021.
Efe, J.A., Hilcove, S., Kim, J., Zhou, H., Ouyang, K., Wang, G., Chen, J., and Ding, S. (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nature cell biology 13, 215–222.
Feng, B., Ng, J.H., Heng, J.C., and Ng, H.H. (2009). Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell stem cell 4, 301–312.
Feng, R., Desbordes, S.C., Xie, H., Tillo, E.S., Pixley, F., Stanley, E.R., and Graf, T. (2008). PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proceedings of the National Academy of Sciences of the United States of America 105, 6057–6062.
Fishman, V.S., Shnayder, T.A., Orishchenko, K.E., Bader, M., Alenina, N., and Serov, O.L. (2015). Cell Divisions are not Essential for the Direct Conversion of Fibroblasts into Neuronal Cells. Cell cycle (Georgetown, Tex), 0.
Forsberg, M., Carlen, M., Meletis, K., Yeung, M.S., Barnabe-Heider, F., Persson, M.A., Aarum, J., and Frisen, J. (2010). Efficient reprogramming of adult neural stem cells to monocytes by ectopic expression of a single gene. Proceedings of the National Academy of Sciences of the United States of America 107, 14657–14661.
Freytag, S.O., Paielli, D.L., and Gilbert, J.D. (1994). Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes & development 8, 1654–1663.
Fusaki, N., Ban, H., Nishiyama, A., Saeki, K., and Hasegawa, M. (2009). Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy Series B, Physical and biological sciences 85, 348–362.
Ginsberg, M., James, D., Ding, B.S., Nolan, D., Geng, F., Butler, J.M., Schachterle, W., Pulijaal, V.R., Mathew, S., Chasen, S.T., et al. (2012). Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFbeta suppression. Cell 151, 559–575.
Gonzalez, F., Barragan Monasterio, M., Tiscornia, G., Montserrat Pulido, N., Vassena, R., Batlle Morera, L., Rodriguez Piza, I., and Izpisua Belmonte, J.C. (2009). Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America 106, 8918–8922.
Guo, Z., Zhang, L., Wu, Z., Chen, Y., Wang, F., and Chen, G. (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell stem cell 14, 188–202.
Gurdon, J.B. (1962). Adult frogs derived from the nuclei of single somatic cells. Developmental biology 4, 256–273.
Han, D.W., Tapia, N., Hermann, A., Hemmer, K., Hoing, S., Arauzo- Bravo, M.J., Zaehres, H., Wu, G., Frank, S., Moritz, S., et al. (2012). Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell stem cell 10, 465–472.
Han, J.K., Chang, S.H., Cho, H.J., Choi, S.B., Ahn, H.S., Lee, J., Jeong, H., Youn, S.W., Lee, H.J., Kwon, Y.W., et al. (2014). Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation 130, 1168–1178.
Heinrich, C., Blum, R., Gascon, S., Masserdotti, G., Tripathi, P., Sanchez, R., Tiedt, S., Schroeder, T., Gotz, M., and Berninger, B. (2010). Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS biology 8, e100037–3.
Heinrich, C., and Rouaux, C. (2015). [Inducing brain regeneration from within: in vivo reprogramming of endogenous somatic cells into neurons]. Medecine sciences : M/S 31, 35–42.
Heinrich, E.M., and Dimmeler, S. (2012). MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circulation research 110, 1014–1022.
Hockemeyer, D., Soldner, F., Beard, C., Gao, Q., Mitalipova, M., DeKelver, R.C., Katibah, G.E., Amora, R., Boydston, E.A., Zeitler, B., et al. (2009). Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature biotechnology 27, 851–857.
Horii, T., Morita, S., Kimura, M., Kobayashi, R., Tamura, D., Takahashi, R.U., Kimura, H., Suetake, I., Ohata, H., Okamoto, K., et al. (2013). Genome engineering of mammalian haploid embryonic stem cells using the Cas9/RNA system. PeerJ 1, e23–0.
Hou, P., Li, Y., Zhang, X., Liu, C., Guan, J., Li, H., Zhao, T., Ye, J., Yang, W., Liu, K., et al. (2013). Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science (New York, NY) 341, 651–654.
Hu, E., Tontonoz, P., and Spiegelman, B.M. (1995). Transdifferentiation of myoblasts by the adipogenic transcription factors PPAR gamma and C/EBP alpha. Proceedings of the National Academy of Sciences of the United States of America 92, 9856–9860.
Huang, P., He, Z., Ji, S., Sun, H., Xiang, D., Liu, C., Hu, Y., Wang, X., and Hui, L. (2011). Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389.
Huang, X., Wang, Y., Yan, W., Smith, C., Ye, Z., Wang, J., Gao, Y., Mendelsohn, L., and Cheng, L. (2015). Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem cells (Dayton, Ohio).
Ieda, M., Fu, J.D., Delgado-Olguin, P., Vedantham, V., Hayashi, Y., Bruneau, B.G., and Srivastava, D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386.
Jaenisch, R., and Young, R. (2008). Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582.
Jayawardena, T.M., Finch, E.A., Zhang, L., Zhang, H., Hodgkinson, C.P., Pratt, R.E., Rosenberg, P.B., Mirotsou, M., and Dzau, V.J. (2015). MicroRNA Induced Cardiac Reprogramming In Vivo: Evidence for Mature Cardiac Myocytes and Improved Cardiac Function. Circulation research 116, 418–424.
Jia, F., Wilson, K.D., Sun, N., Gupta, D.M., Huang, M., Li, Z., Panetta, N.J., Chen, Z.Y., Robbins, R.C., Kay, M.A., et al. (2010). A nonviral minicircle vector for deriving human iPS cells. Nature methods 7, 197–199.
Karamariti, E., Margariti, A., Winkler, B., Wang, X., Hong, X., Baban, D., Ragoussis, J., Huang, Y., Han, J.D., Wong, M.M., et al. (2013). Smooth muscle cells differentiated from reprogrammed embryonic lung fibroblasts through DKK3 signaling are potent for tissue engineering of vascular grafts. Circulation research 112, 1433–1443.
Kim, D., Kim, C.H., Moon, J.I., Chung, Y.G., Chang, M.Y., Han, B.S., Ko, S., Yang, E., Cha, K.Y., Lanza, R., et al. (2009). Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell stem cell 4, 472–476.
Kim, J., Su, S.C., Wang, H., Cheng, A.W., Cassady, J.P., Lodato, M.A., Lengner, C.J., Chung, C.Y., Dawlaty, M.M., Tsai, L.H., et al. (2011). Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell stem cell 9, 413–419.
Kim, Y.J., Lim, H., Li, Z., Oh, Y., Kovlyagina, I., Choi, I.Y., Dong, X., and Lee, G. (2014). Generation of multipotent induced neural crest by direct reprogramming of human postnatal fibroblasts with a single transcription factor. Cell stem cell 15, 497–506.
King, T.J., and Briggs, R. (1955). Changes in the nuclei of differentiating gastrula cells, as demonstrated by nuclear transplantation. Proceedings of the National Academy of Sciences of the United States of America 41, 321–325.
Kretsovali, A., Hadjimichael, C., and Charmpilas, N. (2012). Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem cells international 2012, 184154.
Kulessa, H., Frampton, J., and Graf, T. (1995). GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes & development 9, 1250–1262.
Kurian, L., Sancho-Martinez, I., Nivet, E., Aguirre, A., Moon, K., Pendaries, C., Volle-Challier, C., Bono, F., Herbert, J.M., Pulecio, J., et al. (2013). Conversion of human fibroblasts to angioblast-like progenitor cells. Nature methods 10, 77–83.
Laiosa, C.V., Stadtfeld, M., Xie, H., de Andres-Aguayo, L., and Graf, T. (2006). Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity 25, 731–744.
Lassar, A.B., Thayer, M.J., Overell, R.W., and Weintraub, H. (1989). Transformation by activated ras or fos prevents myogenesis by inhibiting expression of MyoD1. Cell 58, 659–667.
Lee, J., Sayed, N., Hunter, A., Au, K.F., Wong, W.H., Mocarski, E.S., Pera, R.R., Yakubov, E., and Cooke, J.P. (2012). Activation of innate immunity is required for efficient nuclear reprogramming. Cell 151, 547–558.
Li, J., Huang, N.F., Zou, J., Laurent, T.J., Lee, J.C., Okogbaa, J., Cooke, J.P., and Ding, S. (2013a). Conversion of human fibroblasts to functional endothelial cells by defined factors. Arteriosclerosis, thrombosis, and vascular biology 33, 1366–1375.
Li, K., Zhu, S., Russ, H.A., Xu, S., Xu, T., Zhang, Y., Ma, T., Hebrok, M., and Ding, S. (2014). Small molecules facilitate the reprogramming of mouse fibroblasts into pancreatic lineages. Cell stem cell 14, 228–236.
Li, W., Li, K., Wei, W., and Ding, S. (2013b). Chemical approaches to stem cell biology and therapeutics. Cell stem cell 13, 270–283.
Loh, K.M., and Lim, B. (2011). A precarious balance: pluripotency factors as lineage specifiers. Cell stem cell 8, 363–369.
Lu, J., Dong, H., Lin, L., Wang, Q., Huang, L., and Tan, J. (2014). miRNA-302 facilitates reprogramming of human adult hepatocytes into pancreatic islets-like cells in combination with a chemical defined media. Biochemical and biophysical research communications 453, 405–410.
Lu, J., Zhang, F., and Kay, M.A. (2013). A mini-intronic plasmid (MIP): a novel robust transgene expression vector in vivo and in vitro. Molecular therapy : the journal of the American Society of Gene Therapy 21, 954–963.
Ma, T., Xie, M., Laurent, T., and Ding, S. (2013). Progress in the reprogramming of somatic cells. Circulation research 112, 562–574.
Marro, S., and Yang, N. (2014). Transdifferentiation of mouse fibroblasts and hepatocytes to functional neurons. Methods in molecular biology (Clifton, NJ) 1150, 237–246.
Najm, F.J., Lager, A.M., Zaremba, A., Wyatt, K., Caprariello, A.V., Factor, D.C., Karl, R.T., Maeda, T., Miller, R.H., and Tesar, P.J. (2013). Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nature biotechnology 31, 426–433.
Neo, W.H., Yap, K., Lee, S.H., Looi, L.S., Khandelia, P., Neo, S.X., Makeyev, E.V., and Su, I.H. (2014). MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. The Journal of biological chemistry 289, 20788–20801.
Okita, K., Nakagawa, M., Hyenjong, H., Ichisaka, T., and Yamanaka, S. (2008). Generation of mouse induced pluripotent stem cells without viral vectors. Science (New York, NY) 322, 949–953.
Parent, J.M., and Anderson, S.A. (2015). Reprogramming patientderived cells to study the epilepsies. Nature neuroscience 18, 360–366.
Patel, T., Tursun, B., Rahe, D.P., and Hobert, O. (2012). Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell reports 2, 1178–1186.
Pfisterer, U., Wood, J., Nihlberg, K., Hallgren, O., Bjermer, L., Westergren-Thorsson, G., Lindvall, O., and Parmar, M. (2011). Efficient induction of functional neurons from adult human fibroblasts. Cell cycle (Georgetown, Tex) 10, 3311–3316.
Riddell, J., Gazit, R., Garrison, B.S., Guo, G., Saadatpour, A., Mandal, P.K., Ebina, W., Volchkov, P., Yuan, G.C., Orkin, S.H., et al. (2014). Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157, 549–564.
Sandler, V.M., Lis, R., Liu, Y., Kedem, A., James, D., Elemento, O., Butler, J.M., Scandura, J.M., and Rafii, S. (2014). Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature 511, 312–318.
Sayed, N., Wong, W.T., Ospino, F., Meng, S., Lee, J., Jha, A., Dexheimer, P., Aronow, B.J., and Cooke, J.P. (2015). Transdifferentiation of human fibroblasts to endothelial cells: role of innate immunity. Circulation 131, 300–309.
Schindeler, A., Yu, N.Y.C., Cheng, T.L., Sullivan, K., Mikulec, K., Peacock, L., Matthews, R., and Little, D.G. (2015). Local Delivery of the Cationic Steroid Antibiotic CSA-90 Enables Osseous Union in a Rat Open Fracture Model of Staphylococcus aureus Infection. J Bone Joint Surg Am 97, 302–309.
Sebastiano, V., Zhen, H.H., Haddad, B., Bashkirova, E., Melo, S.P., Wang, P., Leung, T.L., Siprashvili, Z., Tichy, A., Li, J., et al. (2014). Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Science translational medicine 6, 264ra163.
Sekiya, S., and Suzuki, A. (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393.
Simeonov, K.P., and Uppal, H. (2014). Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PloS one 9, e10013–4.
Stadtfeld, M., and Hochedlinger, K. (2010). Induced pluripotency: history, mechanisms, and applications. Genes & development 24, 2239–2263.
Stadtfeld, M., Nagaya, M., Utikal, J., Weir, G., and Hochedlinger, K. (2008). Induced pluripotent stem cells generated without viral integration. Science (New York, NY) 322, 945–949.
Takahashi, K., Okita, K., Nakagawa, M., and Yamanaka, S. (2007a). Induction of pluripotent stem cells from fibroblast cultures. Nature protocols 2, 3081–3089.
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., and Yamanaka, S. (2007b). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872.
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676.
Takeuchi, J.K., and Bruneau, B.G. (2009). Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708–711.
Thier, M., Worsdorfer, P., Lakes, Y.B., Gorris, R., Herms, S., Opitz, T., Seiferling, D., Quandel, T., Hoffmann, P., Nothen, M.M., et al. (2012). Direct conversion of fibroblasts into stably expandable neural stem cells. Cell stem cell 10, 473–479.
Tontonoz, P., Hu, E., and Spiegelman, B.M. (1994). Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79, 1147–1156.
Vierbuchen, T., Ostermeier, A., Pang, Z.P., Kokubu, Y., Sudhof, T.C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041.
Wang, L., Wang, L., Huang, W., Su, H., Xue, Y., Su, Z., Liao, B., Wang, H., Bao, X., Qin, D., et al. (2013). Generation of integration-free neural progenitor cells from cells in human urine. Nature methods 10, 84–89.
Wapinski, O.L., Vierbuchen, T., Qu, K., Lee, Q.Y., Chanda, S., Fuentes, D.R., Giresi, P.G., Ng, Y.H., Marro, S., Neff, N.F., et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635.
Warren, L., Manos, P.D., Ahfeldt, T., Loh, Y.H., Li, H., Lau, F., Ebina, W., Mandal, P.K., Smith, Z.D., Meissner, A., et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell stem cell 7, 618–630.
Warren, L., Ni, Y., Wang, J., and Guo, X. (2012). Feeder-free derivation of human induced pluripotent stem cells with messenger RNA. Scientific reports 2, 65–7.
Weider, M., Wegener, A., Schmitt, C., Kuspert, M., Hillgartner, S., Bosl, M.R., Hermans-Borgmeyer, I., Nait-Oumesmar, B., and Wegner, M. (2015). Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells. PLoS genetics 11, e100500–8.
Wiley, L.A., Burnight, E.R., Songstad, A.E., Drack, A.V., Mullins, R.F., Stone, E.M., and Tucker, B.A. (2015). Patient-specific induced pluripotent stem cells (iPSCs) for the study and treatment of retinal degenerative diseases. Progress in retinal and eye research 44, 15–35.
Wilmut, I., Schnieke, A.E., McWhir, J., Kind, A.J., and Campbell, K.H. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813.
Xia, G., Gao, Y., Jin, S., Subramony, S., Terada, N., Ranum, L.P., Swanson, M.S., and Ashizawa, T. (2015). Genome Modification Leads to Phenotype Reversal in Human Myotonic Dystrophy type 1 iPS-cell Derived Neural Stem Cells. Stem cells (Dayton, Ohio).
Xie, H., Ye, M., Feng, R., and Graf, T. (2004). Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676.
Yang, R., Zheng, Y., Li, L., Liu, S., Burrows, M., Wei, Z., Nace, A., Herlyn, M., Cui, R., Guo, W., et al. (2014). Direct conversion of mouse and human fibroblasts to functional melanocytes by defined factors. Nature communications 5, 580–7.
Yoo, A.S., Sun, A.X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., Lee-Messer, C., Dolmetsch, R.E., Tsien, R.W., and Crabtree, G.R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231.
Yoshioka, N., Gros, E., Li, H.R., Kumar, S., Deacon, D.C., Maron, C., Muotri, A.R., Chi, N.C., Fu, X.D., Yu, B.D., et al. (2013). Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell stem cell 13, 246–254.
Yu, B., He, Z.Y., You, P., Han, Q.W., Xiang, D., Chen, F., Wang, M.J., Liu, C.C., Lin, X.W., Borjigin, U., et al. (2013). Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell stem cell 13, 328–340.
Yu, J., Vodyanik, M.A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J.L., Tian, S., Nie, J., Jonsdottir, G.A., Ruotti, V., Stewart, R., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 318, 1917–1920.
Yuan, Y., Hartland, K., Boskovic, Z., Wang, Y., Walpita, D., Lysy, P.A., Zhong, C., Young, D.W., Kim, Y.K., Tolliday, N.J., et al. (2013). A small-molecule inducer of PDX1 expression identified by highthroughput screening. Chemistry & biology 20, 1513–1522.
Zhou, H., Wu, S., Joo, J.Y., Zhu, S., Han, D.W., Lin, T., Trauger, S., Bien, G., Yao, S., Zhu, Y., et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell stem cell 4, 381–384.
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J., and Melton, D.A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to betacells. Nature 455, 627–632.
Zhu, S., Rezvani, M., Harbell, J., Mattis, A.N., Wolfe, A.R., Benet, L.Z., Willenbring, H., and Ding, S. (2014). Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature 508, 93–97.
Author information
Authors and Affiliations
Corresponding author
Additional information
Open Access
This article is distributed under the terms of the Creative Commons Attribution License (CC-BY 4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited..
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Van Pham, P. Direct reprogramming of somatic cells: an update. Biomed Res Ther 2, 8 (2015). https://doi.org/10.7603/s40730-015-0008-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.7603/s40730-015-0008-y