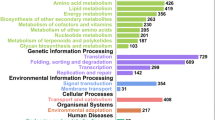
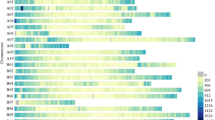
Abstract
Capsicum chinense, is closely related to Capsicum annuum L., and the two species share a common gene pool. There are a limited number of cultivated C. chinense varieties; yet, it is acknowledged that C. chinense germplasm harbors useful alleles for important traits such as fruit yield, disease resistance, accumulation of secondary metabolites and technological fruit characters. Besides, it is feasible to obtain fertile progeny from interspecific crosses of C. chinense and C. annuum. Consequently, C. chinense has high breeding potential for improvement of several different traits in pepper breeding. In the present work, C. chinense genome assembly was mined for trinucleotide simple sequence repeats (SSRs) and repeat loci were converted to 53,749 PCR markers. Marker transferability to C. annuum genome was analyzed with high-stringency cross-species mapping parameters, identifying 17,992 transferable markers. Selecting for interspecific polymorphisms and eliminating redundant loci resulted in 4994 SSR markers distributed over the 12 pepper chromosomes with high potential for interspecific polymorphisms between the two pepper species. Laboratory experiments with a subset of candidate polymorphic markers (36 markers) resulted in an amplification rate of 100% from both pepper species and 31 markers produced polymorphic alleles. Thus, in silico marker development and cross-species mapping analyses proved highly efficient in accuracy of primer design and detection of interspecific sequence divergence. The large set of genome-anchored SSR markers introduced in the present work has high potential for polymorphisms. Therefore, the marker set constitutes a useful genomic resource for broadening the genetic base of cultivated pepper with favorable C. chinense alleles.


Similar content being viewed by others
References
Aguilar-Melѐndez A, Morrell PL, Roose ML, Kim SC (2009) Genetic diversity and structure in semiwild and domesticated chiles (Capsicum annuum; Solanaceae) from Mexico. Am J Bot 96:1190–1202. https://doi.org/10.3732/ajb.0800155
Berke TG, Shieh SC (2012) Capsicum cultivars. In: Peter KV (ed) Handbook of Herbs and Spices, 2nd edn. Woodhead Publishing, Cambridge, pp 116–130. https://doi.org/10.1533/9780857095671.116
Bhattarai G, Mehlenbacher SA (2017) In silico development and characterization of tri nucleotide simple sequence repeat markers in hazelnut (Corylusavellana L.). PLoS ONE 12:e0178061. https://doi.org/10.1371/journal.pone.0178061
Boiteux LS, Nagata T, Dutra WP, Fonseca MEN (1993) Sources of resistance to tomato spotted wilt virus (TSWV) in cultivated and wild species of Capsicum. Euphytica 67:89–94. https://doi.org/10.1007/BF00022729
Boiteux LS, de Ávila AC (1994) Inheritance of a resistance specific to tomato spotted wilt tospovirus in Capsicum chinense ‘PI 159236’. Euphytica 75:139–142. https://doi.org/10.1007/BF00024541
Boiteux LS (1995) Allelic relationships between genes for resistance to tomato spotted wilt tospovirus in Capsicum chinense. Theor Appl Genet 90:146–149. https://doi.org/10.1007/BF00221009
Boukema IW (1979) Allelism of genes controlling resistance to TMV in Capsicum L. Euphytica 29:433–439. https://doi.org/10.1007/BF00025143
Cardenas-Manríquez G, Vega-Muñoz I, Villagómez-Aranda AL, León-Galvan MF, Cruz-Hernandez A, Torres-Pacheco I et al (2016) Proteomic and metabolomics profiles in transgenic tobacco (N. tabacumxanthinc) to CchGLP from Capsicum chinense BG-3821 resistant to biotic and abiotic stresses. Environ Exp Bot 130:33–41. https://doi.org/10.1016/j.envexpbot.2016.05.005
Celik I, Gurbuz N, Uncu AT, Frary A, Doganlar S (2017) Genome-wide SNP discovery and QTL mapping for fruit quality traits in inbred backcross lines (IBLs) of Solanum pimpinellifolium using genotyping by sequencing. BMC Genomics 18:1. https://doi.org/10.1186/s12864-016-3406-7
Cheng J, Zhao Z, Li B, Qin C, Wu Z, Trejo-Saavedra DL et al (2016) A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci Rep 6:18919. https://doi.org/10.1038/srep18919
Cui J, Cheng J, Nong D, Peng J, Hu Y, He W et al (2017) Genome-wide analysis of simple sequence repeats in bitter gourd (Momordicacharantia). Front Plant Sci 8:1103. https://doi.org/10.3389/fpls.2017.01103
Dettori MT, Micali S, Giovinazzi J, Scalabrin S, Verde I, Cipriani G (2015) Mining microsatellites in peach genome: development of new long-core SSR markers for genetic analyses in five Prunus species. Springer Plus 4:337. https://doi.org/10.1186/s40064-015-1098-0
Dossa K, Yu J, Liao B, Cisse N, Zhang X (2017) Development of highly informative genome wide single sequence repeat markers for breeding applications in sesame and construction of a web resource: Sisat Base. Front Plant Sci 8:1470. https://doi.org/10.3389/fpls.2017.01470
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
García-Neria MA, Rivera-Bustamante RF (2011) Characterization of Geminivirus resistance in an accession of Capsicum chinense Jacq. Mol Plant-Microbe Interact 24:172–182. https://doi.org/10.1094/MPMI-06-10-0126
Hirsch CN, Hirsch CD, Felcher K, Coombs J, Zarka D, Van Deynze A et al (2013) Retrospective view of North American potato (Solanum tuberosum L.) breeding in the 20th and 21st centuries. G3 3:1003–1013. https://doi.org/10.1534/g3.113.005595
Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA et al (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–278. https://doi.org/10.1038/ng.2877
Kim S, Park J, Yeom SI, Kim YM, Seo E, Kim KT et al (2017) New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol 18:210. https://doi.org/10.1186/s13059-017-1341-9
Lee GR, Shin MK, Yoon DJ, Kim AR, Yu R, Park NH, Han IS (2013) Topical application of capsaicin reduces visceral adipose fat by affecting adipokine levels in high-fat diet-induced obese mice. Obesity 21:115–122. https://doi.org/10.1002/oby.20246
Leung FW (2014) Capsaicin as an anti-obesity drug. Prog Drug Res 68:171–179. https://doi.org/10.1007/978-3-0348-0828-6_7
Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: Structure, function and evolution. MolBiolEvol 21:991–1007. https://doi.org/10.1093/molbev/msh073
Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z et al (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46:1220–1228. https://doi.org/10.1038/ng.3117
Liu J, Qu J, Hu K, Zhang L, Li J, Wu B et al (2015) Development of genomewide simple sequence repeat fingerprints and highly polymorphic markers in cucumbers based on next generation sequence data. Plant Breeding 134:605–611. https://doi.org/10.1111/pbr.12304
Mahasuk P, Struss D, Mongkolporn O (2016) QTLs for resistance to anthracnose identified in two Capsicum sources. Mol Breeding 36:10. https://doi.org/10.1007/s11032-016-0435-5
Manzur JP, Fita A, Prohens J, Rodríguez-Burruezo A (2015) Successful wide hybridization and introgressing breeding in a diverse set of common peppers (Capsicum annuum) using different cultivated Ají (C. baccatum) accessions as donor parents. PLoS ONE 10:e0144142. https://doi.org/10.1371/journal.pone.0144142
Mc Carty MF, Di Nicolantonio JJ, O’Keefe JH (2015) Capsaicin may have important potential for promoting vascular and metabolic health. Open Heart 2:e000262. https://doi.org/10.1136/openhrt-2015-000262
Mejía-Teniente L, Joaquin-Ramos Ade J, Torres-Pacheco I, Rivera-Bustamante RF, Guevara-Olvera L, Rico-García E, Guevara-Gonzales RG (2015) Silencing of a germin-like protein gene (CchGLP) in geminivirus-resistant pepper (Capsicum chinense Jacq.) BG-3821 increases susceptibility to single and mixed infections by geminiviruses PHYVV and PepGMV. Viruses 7:6141–6151. https://doi.org/10.3390/v7122930
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80. https://doi.org/10.1101/gr.10.1.72
Mimura Y, Inoue T, Minamiyama Y, Kubo N (2012) An SSR-based genetic map of pepper (Capsicum annuum L.) serves as an anchor for the alignment of major pepper maps. Breed Sci 62:93–98. https://doi.org/10.1270/jsbbs.62.93
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200. https://doi.org/10.1038/ng822
Nowaczyk P, Nowaczyk L, Olszewska D (2005) Technological characteristics of the lines selected out from interspecific hybrid Capsicum annuum L. × Capsicum chinense Jacq. Veg Crops Res Bull 62:91–96
Nowaczyk P, Nowaczyk L, Olszewska D, Krupska A (2009) Androgenic response of genotypes selected from Capsicum annuum L. × Capsicum chinense Jacq. Hybrids. Acta Physiol Plant 31:877–879. https://doi.org/10.1007/s11738-009-0315-2
Pakdeevaraporn P, Wasee S, Taylor PWJ, Mongkolporn O (2005) Inheritance of resistance to anthracnose caused by Colletotrichumcapsici in Capsicum. Plant Breeding 124:206–208. https://doi.org/10.1111/j.1439-0523.2004.01065.x
Qin C, Yu C, Shen Y, Fang X, Chen L, Min J et al (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl AcadSci USA 111:5135–5140. https://doi.org/10.1073/pnas.1400975111
Schuler GD (1997) Sequence mapping by electronic PCR. Genome Res 7:541–550. https://doi.org/10.1101/gr.7.5.541
Shirasawa K, Koilkonda P, Aoki K, Hirakawa H, Tabata S, Watanabe M et al (2012) In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol 12:80. https://doi.org/10.1186/1471-2229-12-80
Shirasawa K, Ishii K, Kim C, Ban T, Suzuki M, Ito T et al (2013) Development of Capsicum EST-SSR markers for species identification and in silico mapping onto the tomato genome sequence. Mol Breeding 31:101–110. https://doi.org/10.1007/s11032-012-9774-z
Sugita T, Semi Y, Sawada H, Utoyama Y, Hosomi Y, Yoshimito E et al (2013) Development of simple sequence repeat markers and construction of a high-density linkage map of Capsicum annuum. Mol Breeding 31:909–920. https://doi.org/10.1007/s11032-013-9844-x
Sui YH, Hui NB (2015) Acquisition, identification and analysis of an interspecific Capsicum hybrid (C. annuum × C. chinense). J Hortic Sci Biotechnol 90:31–38. https://doi.org/10.1080/14620316.2015.11513150
Sun CY, Mao SL, Zhang ZH, Palloix A, Wang LH, Zhang BX (2015) Resistance to anthracnose (Colletotrichumacutatum) of Capsicum mature green and ripe fruit are controlled by a major dominant cluster of QTLs on chromosome P5. SciHortic 181:81–88. https://doi.org/10.1016/j.scienta.2014.10.033
Tan S, Cheng JW, Zhang L, Qin C, Nong DG, Li WP et al (2015) Construction of an interspecific genetic map based on InDel and SSR for mapping the QTLs affecting the initiation of flower primordia in pepper (Capsicum spp.). PLoS ONE 10:e0119389. https://doi.org/10.1371/journal.pone.0119389
Tang J, Baldwin SJ, Jacobs JME, van der Linden CG, Voorrips RE, Leunissen JAM et al (2008) Large-scale identification of polymorphic microsatellites using an in silico approach. BMC Bioinformatics 9:374. https://doi.org/10.1186/1471-2105-9-374
Tanksley SD, Iglesias-Olivas J (1984) Inheritance and transfer of multiple-flower character from Capsicum chinense to Capsicum annuum. Euphytica 33:769–777. https://doi.org/10.1007/BF00021903
Uncu AT, Celik I, Devran Z, Ozkaynak E, Frary A, Frary A, Doganlar S (2015) Development of a SNP-based CAPS assay for the Me1 gene conferring resistance to root knot nematode in pepper. Euphytica 206:393–399. https://doi.org/10.1007/s10681-015-1489-x
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Voorrips RE, Finkers R, Sanjaya L, Groenwold R (2004) QTL mapping of anthracnose (Colletotrichum spp.) resistance in a cross between Capsicum annuum and C. chinense. Theor Appl Genet 109:1275–1282. https://doi.org/10.1007/s00122-004-1738-1
Wahyuni Y, Ballester AR, Sudarmonowati E, Bino RJ, Bovy AG (2011) Metabolite biodiversity in pepper (Capsicum) fruits of thirty-two diverse accessions: Variation in health related compounds and implications for breeding. Phytochemistry 72:1358–1370. https://doi.org/10.1016/j.phytochem.2011.03.016
Wang X, Wang L (2016) GMATA: An integrated software package for genome-scale SSR mining, marker development and viewing. Front Plant Sci 7:1350. https://doi.org/10.3389/fpls.2016.01350
Zhang XF, Sun HH, Xu Y, Chen B, Yu SC, Geng SS, Wang Q (2016) Development of a large number of SSR and InDel markers and construction of a high-density genetic map based on a RIL population of pepper (Capsicum annuum L.). Mol Breeding 36:92. https://doi.org/10.1007/s11032-016-0517-4
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Uncu, A.T. Genome-wide identification of simple sequence repeat (SSR) markers in Capsicum chinense Jacq. with high potential for use in pepper introgression breeding. Biologia 74, 119–126 (2019). https://doi.org/10.2478/s11756-018-0155-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11756-018-0155-x