Abstract
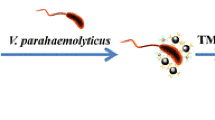
A developed Christmas-tree derived immunosensor based on a gold label silver stain (GLSS) technique was fabricated for a highly sensitive analysis of Vibrio parahaemolyticu (VP). In this strategy, captured VP antibody (cAb) was immobilized on a solid substrate; then, the VPs were sequentially tagged with a signal probe by incubating the assay with a detection VP antibody (dAb) conjugated gold nanoparticles (AuNPs)-labeled graphite-like carbon nitride (g-C3N4). Finally, the attached signal probe could harvest a visible signal by the silver meal deposition, and then followed by homebrew Matlab 6.0 as a grey value acquisition. In addition, the overall design of the biosensor was established in abundant AuNPs and g-C3N4 with a two-dimensional structure, affording a bulb-decorated Christmas-tree model. Moreover, with the optimized conditions, the detection limit of the as-proposed biosensor is as low as 102 CFU (Colony-Forming Units) mL−1, exhibiting an increase of two orders of magnitude compared with the traditional immune-gold method. Additionally, the developed visible immunosensor was also successfully applied to the analysis of complicated samples.
Similar content being viewed by others
References
W. Mala, M. Alam, S. Angkititrakul, S. Wongwajana, V. Lulitanond, S. Huttayananont, and C. Chomvarin, Infec. Genet. Evol., 2016, 39, 212.
M. T. Hossain, Y. O. Kim, and I. S. Kong, Mol. Cell. Probe, 2013, 27, 171.
J. Wei, X. Zhou, D. Xing, and B. Wu, Food Chem., 2010, 123, 852.
B. Bisha, J. Simonson, M. Janes, K. Bauman, and L. D. Goodridge, Int. J. Food Sci. Tech., 2012, 47, 885.
C. C. Li, Z. C. Ye, L. Y. Wen, F. K. Zhao, and J. Y. Pan, Vaccine, 2014, 32, 6115.
Y. Hara-Kudo and S. Kumagai, Epidemiol. Infect., 2014, 142, 2237.
K. S. Shaw, A. R. Sapkota, J. M. Jacobs, and B. C. Crump, Environ. Int., 2015, 74, 99.
O. Laczka, M. Labbate, and M. Doblin, Anal. Methods, 2014, 6, 2020.
W. Jiang, X. Han, Q. Wang, and C. Ding, Appl. Microbiol. Biot., 2014, 98, 4937.
B. K. Kumar, P. Raghunath, D. Devegowda, V. K. Deekshit, M. N. Venugopal, and I. Karunasagar, Int. J. Food Microbiol, 2011, 145, 244.
Y. R. Kim, E. Y. Kim, D. G. Kim, Y. O. Kim, M. T. Hossain, and I. S. Kong, Anal. Biochem., 2012, 424, 32.
E. Sloan, M. O’Neill, C. Kaysner, A. DePaola, J. L. Nordstrom, and J. Sofos, J. Rapid Meth. Aut. Mic., 2003, 11, 297.
N. Miwa, M. Kashiwagi, F. Kawamori, T. Masuda, Y. Sano, M. Hiroi, and H. Kurashige, J. Food Hyg. Soc. Jpn., 2006, 47, 41.
J. L. Jones, Y. Hara-Kudo, J. A. Krantz, R. A. Benner, and A. DePaola, Food Microbiol., 2012, 30, 105.
P. He, Z. Chen, J. Luo, H. Wang, and W. Gao, Mol. Cell Probe, 2014, 28, 246.
F. Zhao, D. Q. Zhou, H. H. Cao, and Y. H. Jiang, Food Control, 2011, 22, 1095.
Z. Zhang, L. Xiao, Y. Lou, M. Jin, and Y. Zhao, Food Control, 2015, 51, 31.
A. Tyagi, V. Saravanan, and I. Karunasagar, Int. J. Food Microbiol, 2009, 129, 124.
G. Xiang, X. Pu, D. Jiang, L. Liu, C. Liu, and X. Liu, PLOS ONE, 2013, 8, e72342.
C. D. Tian, Y. Z. Zhang, X. Y. Ma, and J. Wang, J. Food Safety, 2011, 31, 371.
L. Wang, L. Shi, J. Su, Y. Ye, and Q. Zhong, Gene, 2013, 515, 421.
Q. Zhong, J. Tian, B. Wang, and L. Wang, Food Control, 2016, 63, 230.
Y. Ye, H. Li, Q. Wu, L. Na, and Y. Han, J. Food Safety, 2014, 34, 21.
Y. Sha, X. Zhang, W. Li, W. Wu, S. Wang, Z. Guo, J. Zhou, and X. Su, Talanta, 2016, 147, 220.
H. C. Chen and T. C. Chang, Int. J. Food Microbiol., 1996, 29, 311.
S. R. Ahmed, S. Oh, R. Baba, H. Zhou, S. Hwang, J. Lee, and E. Y. Park, Nanoscale Res. Lett., 2016, 11, 1.
T. Ding, A. W. Rudrum, L. O. Herrmann, V. Turek, and J. J. Baumberg, Nanoscale, 2016, 8, 15864.
Ł. Lentka, M. Kotarski, J. Smulko, U. Cindemir, Z. Topalian, C. G. Granqvist, R. Calavia, and R. Lonescu, Talanta, 2016, 160, 9.
L. Nie, J. Tang, H. Guo, H. Chen, P. Xiao, and N. He, Anal. Sci., 2004, 20, 461.
R. Q. Liang, C. Y. Tan, and K. C. Ruan, J. Immunol. Methods, 2004, 285, 157.
H. J. Qi, S. H. Chen, M. L. Zhang, H. Shi, and S. Q. Wang, Anal. Bioanal. Chem., 2010, 398, 2745.
Y. Deng, L. Tang, G. Zeng, J. Wang, Y. Zhou, J. Wang, J. Tang, Y. Liu, B. Peng, and F. Chen, J. Mol. Catal. A, 2016, 421, 209.
Z. Huang, F. Li, B. Chen, and G. Yuan, RSC Adv., 2015, 5, 14027.
E. Z. Lee, S. U. Lee, N. S. Heo, G. D. Stucky, Y. S. Jun, and W. H. Hong, Chem. Commun., 2012, 48, 3942.
Y. Bai, P. Q. Wang, J. Y. Liu, and X. J. Liu, RSC Adv., 2014, 4, 19456.
X. Li, Z. Guo, J. Li, Y. Zhang, H. Ma, X. Pang, B. Du, and Q. Wei, Anal. Chim. Acta, 2015, 854, 40.
Q. Liu, Y. Guo, Z. Chen, Z. Zhang, and X. Fang, Appl. Catal. B, 2016, 183, 231.
S. Chen, A. Li, L. Zhang, and J. Gong, Anal. Chim. Acta, 2015, 896, 68.
L. Du, Y. Wang, Z. Ren, C. Shen, and G. Luo, Ind. Eng. Chem. Res., 2016, 55, 6783.
L. Chen, X. Zeng, P. Si, Y. Chen, Y. Chi, D. H. Kim, and G. Chen, Anal. Chem., 2014, 86, 4188.
Z. Zhang, C. Chen, and X. S. Zhao, Electroanalysis, 2009, 21, 1316.
W. Wang, W. Y. Wu, X. Zhong, W. Wang, Q. Miao, and J. J. Zhu, Biosens. Bioelectron., 2011, 26, 3110.
Acknowledgments
Financial support from the National Natural Science Foundation of China (41576098, 81273130, and 511601370), the Science and Technology Department of Zhejiang Province of China (2016C33176, 2016R405068), the Natural Sciences Foundation of Guangdong Province of China (2014A030313590) and Shenzhen Science and Technology Bureau (JCYJ20140508155916417) are gratefully acknowledged. This work is also sponsored by K. C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Song, X., Wu, Y., Wu, L. et al. Christmas-tree Derived Amplification Immuno-Strategy for Sensitive Visual Detection of Vibrio parahaemolyticus Based on Gold Label Silver Stain Technology. ANAL. SCI. 33, 889–895 (2017). https://doi.org/10.2116/analsci.33.889
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2116/analsci.33.889