Abstract
There is a paradigm shift towards data-centric AI, where model efficacy relies on quality, unified data. The common research analytics and data lifecycle environment (CRADLE™) is an infrastructure and framework that supports a data-centric paradigm and materials data science at scale through heterogeneous data management, elastic scaling, and accessible interfaces. We demonstrate CRADLE’s capabilities through five materials science studies: phase identification in X-ray diffraction, defect segmentation in X-ray computed tomography, polymer crystallization analysis in atomic force microscopy, feature extraction from additive manufacturing, and geospatial data fusion. CRADLE catalyzes scalable, reproducible insights to transform how data is captured, stored, and analyzed.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Modern materials science contends with immense volumes of heterogeneous data from experiments and simulations. High powered characterization techniques, for example, can capture data at rates up to 30 GB/sec.[[1]] This necessitates paradigm shift in the methodology of materials science and data pipelines that can handle information at scale.[[2,3]] Large-scale high performance computing (HPC), artificial intelligence (AI), and machine learning (ML) have each become integral tools to generate scientific insights from massive datasets. This follows a broader trend in the scientific community towards AI4Science, utilizing the power of AI/ML to enable scientific discovery.[[4,5]]
The integration of AI/ML in materials science, however, introduces several key challenges. First, the scale of modern datasets generated during experimentation requires now domain scientists to possess expertise in software and data engineering to generate insights. This demand has resulted in the generation of a new role: the materials data scientist. Second, the scale and heterogeneity of materials data requires a complex computational infrastructure to manage it. Existing Big Data infrastructures, however, are often designed by technology companies who have different needs from scientific research. This misalignment means current systems and frameworks fail to support the domain needs of materials data science. Furthermore, modern research tends to focus on the improvement of models and algorithms over the data itself. The growing field of data-centric AI pushes against this trend, advocating for more systematic practices. In data-centric AI, datasets are treated as fluid entities that evolve alongside models to avoid the “garbage in = garbage out” problem.[[6,7,8]]
The common research analytics and data lifecycle environment (CRADLE) was developed as a comprehensive framework to address this gap and support data-centric AI.[[9,10,11]] CRADLE provides a computing infrastructure and framework that enables materials data science at scale. Key components includes multimodal data processing, accessible interfaces for complex computational engines, interactive visualization tools, and distributed computing storage. In this work, we demonstrate how CRADLE enables novel materials science studies on large-scale datasets across different modalities. Examples include distributed data ingestion and accelerated deep learning for phase identification in X-ray diffraction (XRD), segmentation of defects in X-ray computed tomography (XCT), analysis of crystallization kinetics in polymers from atomic force microscopy (AFM) imaging, feature extraction from additive manufacturing (AM), and integrating satellite imagery for monitoring crop health.
Background
Distributed and High-Performance Computing
High performance computing (HPC) systems consist of tightly integrated clusters of servers that incorporate specialized hardware to achieve exceptional computing power. These systems are composed of specialized hardware such as high-core count CPUs, accelerators like GPUs, and parallel storage systems.[[12]] This hardware and design enables them to run complex simulations, train machine learning models, and perform advanced scientific calculations. These tasks benefit from the tightly coupled nature of HPC clusters, where high-speed networking interconnects ensure efficient communication and data exchange between nodes. HPC systems are designed for vertical scaling, which involves augmenting existing nodes with more powerful resources like additional cores, higher memory capacities, and faster input/output (I/O) capabilities.
Distributed systems consist of a network of commodity hardware, that utilizes the collective power of many servers to process and store Big Data. These systems leverage frameworks like Hadoop to process large datasets across many standard computers.[[13]] Tasks are split into smaller subtasks, distributed to nodes for processing, and then aggregated, enabling the system to analyze and process vast amounts of data efficiently.[[14]] These systems are designed to be highly fault-tolerant, automatically handling failures without disrupting the overall operation. Distributed computing systems are designed to scale horizontally, which involves the addition of more nodes to increase processing capability and storage capacity. This allows for cost-effective scaling, as it relies on standard, off-the-shelf hardware rather than specialized, high-cost components.
Materials Data Science Infrastructure
Much of the development in large-scale data infrastructure has been pioneered by Internet companies. Companies such as Airbnb, LinkedIn and Uber receive massive data from millions of users, and have accordingly developed both scale-up and scale-out infrastructure to store, process and generate insights from this data.[[15,16]] These organizations, however, have fundamentally different design interests than research institutions and experimental scientists. As a result, numerous research groups have developed custom data infrastructures and components for materials data.
Experimental data pipelines are designed to manage the flow of big data from the instruments that collect it to a storage location. This requires tight integration with instruments and low-latency networking. NREL’s research data infrastructure (RDI) gathers data from numerous instruments across the campus and writes information to a PostgreSQL database for analysis.[[17]] Kadi4Mat provides researchers with an electronic lab notebook (ELN) environment that lets users interact and program with data as it is collected from instruments.[[18]]
Designing data standards and databases is another key research area to catalog existing materials and discover new materials. Numerous efforts have been made to develop such these catalogs, such as the open quantum materials database (OQMD) and Automatic-FLOW for materials discovery (AFLOW).[[19,20]] There has been a particular emphasis on building data with FAIR principles; making it Findable, Accessible, Interoperable, and Reusable.[[21,22]]
Machine learning pipelines have also been developed to generate robust predictive models from large datasets. Examples include data from tomography beamlines, as well as from X-ray ptychography.[[23,24]] Some of these experiments incorporate a control pipeline, allowing automated, real-time responses to failures in the experimental pipeline.[[25,26]]
Required Capabilities for Data-Centric AI
Existing materials data science infrastructure fails to fully meet emerging needs. Current systems remain siloed by instrumentation types and data modalities, lacking integration to consolidate insights. Support for scalable and reproducible analytics using techniques like machine learning across large, multimodal datasets is limited. As a result, significant software and data engineering expertise is still required alongside scientific domain knowledge. Several key challenges are described below.
Materials data is highly fragmented and heterogeneous, spanning structured tables, images, videos, spectra, and more. Managing and integrating these diverse modalities at scale is challenging. For example, a additive manufacturing experiment can contain multiple sensors that produce tabular, image, and acoustic data all at different time scales. Optimized storage and processing is needed for each data type (e.g. databases for tables, object stores for files), along with abstractions like graphs to connect insights across modalities.
Managing large datasets and leveraging HPC has a high barrier to entry for materials scientists due to the technical expertise required. Infrastructure should lower barriers for domain scientists without extensive coding expertise via intuitive interfaces, modular building blocks, and managed services. This allows researchers to focus insights rather than navigating complex distributed systems. Open source components also enable sharing of data, models, and algorithms.
Reproducibility of studies is challenging due to ever-changing data management practices. Metadata and identifiers are unique to each experiment making connecting historical studies and heterogenous data difficult. Enforcing FAIR principles across the data lifecycle pipelines enables reproducibility of results. This includes careful design of data schemas and capturing raw data, analysis code, and trained models for published studies in accessible repositories.
Designing a system that can handle high-throughput computing for complex simulations and machine learning model training paired with distributed frameworks to process huge datasets efficiently is non-trivial. The system should leverage both vertical (scale-up) and horizontal (scale-out) scaling to accelerate materials data science. A small research group of five individuals to a large national laboratory of thousands should be able to use the same framework and scale it to their specific needs.
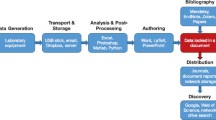
CRADLE provides a holistic solution to address each of these required capabilities and design challenges. Figure 1 depicts the materials data science life cycle and workflow supported by the framework from raw data ingestion to predictive deep learning models. The following section deconstructs the framework’s components and technical details.
CRADLE Framework
Elastic Scaling Architecture
CRADLE utilizes a hybrid infrastructure encompassing a high-performance computing cluster and additional Hadoop clusters to enable both vertical and horizontal scaling. The HPC nodes feature powerful CPUs, accelerators, and high-speed networking to efficiently run simulations, visualizations, and machine learning models. The Hadoop clusters provide a distributed framework over commodity hardware to process huge datasets in parallel.
This architecture supports allocating the right workload to the right system. Small, high-throughput batch jobs that require fast single node performance run on the HPC cluster. Massive jobs needing to process terabytes of data in a fault-tolerant fashion execute on the expandable Hadoop cluster. The systems interface to share data and results, with storage locality minimizing transfer latencies. Proper workload allocation ensures that a user performing data transformations on a million rows of time series data will not monopolize the GPU nodes of a HPC cluster that other users might need for training deep learning models.
A key advantage of this design is cost-effective scalability. Expanding the HPC cluster necessitates purchasing expensive, cutting-edge hardware in a manual process. The Hadoop cluster leverages cheap, off-the-shelf servers and automated workflows to easily grow storage and processing capabilities. CRADLE’s hybrid architecture is composed of the Case Western Reserve University’s HPC cluster alongside a set of three separate Hadoop clusters. The most recent iteration Hadoop cluster contains two petabytes of distributed storage. Full hardware specifications are shown in Fig. 2.
Multimodal Data Storage and Processing
CRADLE’s storage layer leverages a hybrid of Hadoop Distributed File System (HDFS) and HPC Network Attached Storage (NAS) to support diverse data modalities at scale. HDFS provides petabyte-level storage across commodity servers with built-in data replication for fault tolerance. This acts as a long-term data lake. The HPC NAS offers high-performance parallel filesystems for temporary storage and low-latency data access.
The storage ecosystem targets the unique needs of different data types. Columnar data is stored in the HBase non-relational database for efficient, low latency queries. Graph data resides in the JanusGraph network database to enable analytical traversals. The Ozone object store houses images and videos to circumvent the small files problem of HDFS and HBase. Underlying HDFS preserves the complete immutable raw dataset for reproducibility.[[13]]
The processing layer mirrors this heterogeneity support. Custom jobs run on either the Hadoop cluster leveraging YARN resource management or the HPC cluster with SLURM job scheduling.[[27,28]] Distributed data pipelines rely on Spark and its Resilient Distributed Dataset (RDD) abstraction for large-scale batch and streaming tasks.[[29,30]] GraphX provides graph-optimized Spark algorithms.[[31]] Impala is used for SQL queries against tabular data partitioned across nodes.[[32]]
Access control across users utilizes integration with university single sign-on (SSO), Kerberos authentication, and Apache Ranger policies.[[33]] The combination of storage, processing, and security layers enables diverse, large-scale datasets to be manipulated, explored, and shared while maintaining access controls.
Hardware and software stack that composes CRADLE. The top portion depicts the HPC cluster. This includes containerized environments with programming IDEs, OS systems, and resource management tools. The bottom portion depicts the Hadoop cluster. This includes the underlying data storage and processing stack. HDFS and Hbase are used to store tabular/graph data storage and Ozone for image/videos. Respective clusters are securely connected via Kerberos and SSL.
Accessible Containerized Data Science Tooling
CRADLE provides containerized data science tooling that domain scientists can leverage to easily access HPC and Hadoop resources. HPC and Hadoop clusters alone involve complex developer tools to interface with for resources. Access to the SLURM scheduler through the shell and Hadoop through lower-level APIs creates a barrier to entry for non-developers. Furthermore, managing package versions and dependency conflicts in languages such as Python and R generate bottlenecks and inconsistent workflows.
Environment containerization solves these problems. CRADLE leverages Singularity containers to create isolated and reproducible software environments that contain all the relevant tools of a data scientist.[[34]] These containers contain data science programming languages, libraries, dependencies, interactive development environments (IDEs), and other relevant tooling for ML.[[35,36]] Containers are securely run on HPC nodes and are configured to access the Hadoop clusters.
These containers are accessed through a simple, intuitive interface using Open OnDemand.[[37]] Open OnDemand provides browser based access to launch applications such as VSCodium and RStudio. A graphical user interface (GUI) allows a user to select the specific computational resources from HPC such as CPU cores, RAM, and GPUs. After selecting the desired options, a session is launched in broswer to an application that is directly hooked onto the HPC NAS file system. This means that a scientist can leverage HPC and Hadoop resources from anywhere as long as they have Internet access. It enables the ability to scale analysis past the limitations of one’s notebook and work in a consistent, dependency resolved environment.
CRADLE Middleware
CRADLE provides a suite of software tools called “middleware” to further simplify access and usage of the infrastructure. These packages abstract complexity and address common workflow bottlenecks that material scientists face. Middleware is designed so scientists can leverage the powerful tooling of HPC and Hadoop without knowledge past basic scripting in R and Python.
CRADLETools The CRADLETools Python/R package enables directly loading HDFS and HBase data into scripts via intuitive function calls. This avoids the need for users to write Java code or Impala SQL queries just to interface with storage. Custom DataLoaders for PyTorch are also implemented, to enable the ability to stream big datasets from Ozone and HDFS for integrated model training. The DataLoader is able to stream data in real-time without loading the entire dataset locally, while also supporting prefetching and caching of batches of data on the GPU to optimize data throughput.
CRADLEFleets Executing large batches of HPC jobs typically requires intricate SLURM configuration. CRADLEFleets condenses this process down to easy Python/R function calls for allocation, submission, and monitoring. Users simply specify the script, arguments, and resources required while the package handles parallel executions and collating outputs. This enables scientists to perform transformations across thousands of files or train hundreds of deep learning models in parallel on HPC.
CRADLE Data Explorer Exploring available datasets can also pose challenges in navigating stores and learning query languages. Furthermore, historical domain knowledge is required to be aware of what experiments may have been previously conducted and where the results from those experiments sit. CRADLE Data Explorer is a R Shiny application that offers rich visualizations cater to different modalities while simplifying access to materials data lakes. Data stored according to FAIR principles enables a complete historical overview of what exists inside of the multimodal data stores, eliminating the necessity to navigate through specific table names or queries.
Data is visualized according to their specific modality such as: interactive 3D intensity plots of XRD, surface maps of AFM polymer crystallization, and large-scale graphs of Photovoltaic power plants. The R base makes the application easily extensible through the addition of new datasets or visualizations. A complete overview of existing datasets, models, and parameters is offered to accelerate the data wrangling process. Serving as a launchpad for data exploration, scientists can retrieve analysis-ready data and models to focus on conducting research rather than engineering data.
CRADLE-Enabled Materials Data Science
The purpose of CRADLE is to enable materials data science at scale. CRADLE provides a comprehensive computing and infrastructure framework to tackle a diverse set of materials challenges, irrespective of data modality and volume. Table I showcases several exemplar studies CRADLE has enabled.
Large-Scale Ingestion and Phase Identification of 2D X-ray Diffraction Patterns Using Deep Learning
Synchrotron X-ray diffraction (XRD) enables highly detailed characterization of crystalline structure changes. The beamline, however, captures high spatiotemporal resolution studies that produce terabtye-scale datasets with ease. Here, we demonstrate the ability of CRADLE to ingest immense datasets, query metadata, and parallelize training of deep neural networks.[[38,39]] Researchers conducted experiments at the CHESS beamline to produce 21 terabytes of XRD data. A 2D area detector (HEXD) captured 4.5 million 2D diffractograms for downstream analysis. Ingesting and working with the heterogenous data utilizing CRADLE presented solutions at scale. Metadata encoded in Apache Parquet partitioned across the HDFS cluster, enabling low-latency SQL analytics via Impala. 2D diffraction patterns were ingested as TIFF files into Apache Ozone to circumvent small file HDFS issues.
After FAIRification and ingestion, developing machine learning pipelines and predictive models became trivial. A subset of data was selected to train a convolutional neural networks (CNNs) for phase identification. Specifically, CNNs were developed to take a 2D diffractogram as input and predict the corresponding \(\beta\)-phase volume fraction. Leveraging HPC and CRADLEFleets, parallelized hyperparameter tuning of 168 CNN models was performed to achieve a mean squared error (MSE) of 0.0026. CRADLE reduced total training time of the pipeline from 89 h to 1 h compared to conducting analysis on a single HPC node.
Automated Pipelines for Corrosion Detection and Visualization using X-ray Computed Tomography
X-ray computed tomography (XCT) enables structural reliability studies through high resolution imaging of pitting corrosion in metal alloy wires. Quantification of corrosion, however, requires meticulous segmentation of microscopic defects across thousands of XCT slices which is highly time consuming. This barrier severely limits scientific analysis to understand corrosion kinetics. Here we demonstrate CRADLE’s capabilities to support automated pipelines for feature extraction, volumetric reconstruction, and topological transformations.[[40]]
A pitting corrosion study was conducted where Al wire was exposed to salt water droplets. During the experiment, 88 3D scans were taken to monitor material changes over time. These 88 3D scans translated to 87,648 2D cross sectional slices in the form of TIFF images. Images were ingested in CRADLE’s Ozone store for object storage.
An automated pipeline was then developed to perform segmentation on 2D images, reconstruct pits in 3D, and provide a complete statistical characterization of all corrosion as shown in Fig. 3. For segmentation, a U-Net was trained on a small subset of images to identify pits in 2D cross sectional slices.[[41]] The model achieved a precision of 0.88 and recall of 0.90 for the binary segmentation task. The trained model was then used for inference on the entire 87,648 TIFF image dataset, parallelizing predictions across HPC using CRADLEFleets. Volumetric reconstructions, statistical quantification, and topological transforms were also parallelized, reducing complete characterization pipeline from hours to under 30 min. Results and reconstructed volumes represented as sparse matrices were ingested into HBase for future analysis. The machine learning and image processing toolbox offered by CRADLE with integration into modality specific storage, enabled an automated pipleline to quickly be developed and deployed.
Graph Neural Networks to Study Crystallization Kinetics and Similarity in AFM Image Sequences
Atomic force microscopy (AFM) enables nanoscale imaging of phase transitions in materials over time. The characterization technique enables studies on crystallite formation in fluoroelastomer films, producing videos comprising thousands of frames. Manually analyzing each image with a traditional software such as Gwyddion proves prohibitive, requiring months of effort without direct insights. Here, we demonstrate CRADLE’s ability to enable distributed data preprocessing, multimodal storage, and acceleration of deep learning.[[42]]
Thirty-six AFM videos were taken of polymer crystallization under different experimental conditions, generating over 21,317 image frames (comprising roughly 90 GB of data). Experimental data was saved in a machine specific format: IGOR Pro binary wave files (IBW). Spark enabled distributed preprocessing of image frames and experimental metadata from the IBW. Leveraging distributed preprocessing and ingestion, provided a 18x speedup over traditional processing in HPC. Similar to other use cases described, tabular metadata was ingested into HBase and TIFF images into Ozone. Comprehensive preprocessing and storage enabled two downstream deep learning pipelines to be developed.
Left: crystallite segmentation and spatiotemporal graph construction pipeline. Segmentation masks are generated from raw TIFF images. The segmented crystallites are then represented as nodes in a graph and edges formed based on nearest neighbors. Right: scaling pairwise similarity calculations between crystallites. The x-axis depicts the number of crystallites being used for pairwise calculations and the y-axis the total time taken for all calculations.
The first pipeline involved training CNNs to segment crystallites from image frames and to quantify their metrics.[[42,43]] A U-Net was trained to segment crystallite from amorphous region and after hyperparameter tuning, achieved a intersection over union (IoU) score of 0.95. Segmented crystallites were then constructed into graph representations using nearest neighbor calculations. Crystallites were represented as nodes in the graph and edges based on nearest neighbor distances. These graphs provided a lightweight representation of images where graph mining techniques could then be applied. The pipeline is depicted on the left in Fig. 4. Leveraging GPU accelerated tooling from CRADLE such as RAPIDS’ CuPy library cut post-processing in half. Quantified feature vectors and images were persisted to HBase and Ozone.
The second pipeline involved training graph neural networks (GNNs) to predict similarity between crystallites. To generate similarity values between 409,371 crystallites involved 167 billion calculations. Working memory of HPC was overflooded, but distributed Spark workloads partitioned comparisons to fit in memory. Figure 4 plots the relationship between number of crystallites used for pairwise calcuations and the total time taken. A graph was assembled from the output and ingested into JanusGraph. This enabled efficient training 216 graph neural network variants using subgraph querying. CRADLE reduced end-to-end efforts from days to hours by solving obstacles at each phase of both pipelines.
Feature Extraction and Multimodal Data Integration from High-Speed Camera Video of Additive Manufacturing
Laser powder bed fusion shows promise as a more efficient additive manufacturing technique but runs risks of defects like keyhole pores harming end products. Coupled sensor streams, however, enable monitoring melt pools to identify anomalies and build accurate predictive models. Examples of sensors inclue high-speed cameras (HSC) capture video of the fusion process and pyrometers to obtain spectral data from the radiation. Integration of these sensors introduces a complex multimodal data management problem at scale.[[44,45]]
A dataset of 750 tracks was collected where both in-situ and ex-situ monitoring was performed. Data from four modalities was collected: process parameters of the machine, high-speed camera video, pyrometry measurements, and radiography of samples post build. The 750 high-speed camera videos contained 1000 frames each. For each high-speed camera frame, there are 100 correlated pyrometry measurements. High-speed camera videos were ingested into HDFS for long term storage. Frames from when the laser was powered and features were present, were extracted and stored in Ozone object storage in addition to radiography images. Pyrometry and process measurements were stored in HBase. A design schema was generated to link all four unique datasets at different spatiotemporal resolutions to enable efficient queries for desired results. Figure 5 depicts a general schema and set of data modalities.
After ingestion, subsets of high-speed camera frames with appropriate features were queried to train object detection and segmentation models such as U-Net and You Only Look Once (YOLO).[[41,46]] The best models used to perform inference across all high-speed camera frames, and results written back into CRADLE’s HBase storage. Since the schema was previously designed, extracted features such as melt pool geometry could immediately be linked to other data modalities for downstream analysis. This enabled researchers to tie together information from disparate sensors such as high-speed camera spatter size and radiography defect presence for a holistic study across the entire build plate. Multimodal analysis to this scale requires a infrastructure and framework that handle high volumes of heterogeneous data.
Monitoring Crop Growth Through a Multiscale Geospatial Satellite Imagery Analysis
Remote sensing technology provides a comprehensive means of monitoring crop growth, soil conditions, and hydrological dynamics crucial for promoting sustainable agricultural practices and mitigating environmental impact. Integrating datasets from different time and spatial scales, however, is challenging spatiotemporal problem that requires well-designed frameworks and tooling. CRADLE serves as a perfect infrastructure to support these integrations through its elastic scaling.[[47]]
In this study, datasets such as MODIS Aqua satellite imagery, USDA historical crop planting data, the Aster Global Digital Elevation Model (GDEM), and USGS stream gauge readings were downloaded. Datasets contained different spatial resolutions such as MODIS at 250 ms and USDA historical crop planting data at 30 ms. Additionally, different datasets contained varying temporal resolutions such as stream gauge readings from USGS at 15 min intervals and MODIS at a 24 h interval for spectral bands. Sensor readings were written into Parquet files and ingested into HDFS for partitioned storage. GeoTIFFs were ingested into Ozone as objects with each GeoTIFF containing close to 13 million points.
A multimodal workflow was developed to combine these disparate datasets across spatiotemporal resolutions to study complex interplay between different monitoring techniques within Ohio. Crop health was be monitored through spectral band information and correlated to soil nitrogren content to capture the relationship between soli nitrogen availability and crop yields. CRADLE provided a flexible interface to store, query, and transform billions of geospatial data points for this study.
Discussion
Data-Centric AI Infrastructure
The paradigm shift towards data-centric AI is essential in unlocking the full potential of machine learning models.[[6,7]] Traditionally, the focus has been predominantly on refining algorithms and model architectures. However, the efficacy of these models is fundamentally reliant on the quality and availability of data. CRADLE exemplifies this shift by establishing an integrated infrastructure that prioritizes data management and accessibility, thus enabling AI and ML applications to operate at unprecedented scales.
This infrastructure not only supports the integration of disparate datasets through multimodal data ingestion but also employs optimized storage formats to improve data accessibility. Embedded data exploration tools within CRADLE provide deep insights into existing datasets, promoting a proactive approach to computational analysis that integrates the data lifecycle from the very beginning of the research process. Lowering the barrier to entry for domain scientists to leverage HPC and distributed computing democratizes the use of these resources and accelerates scientific research.
Advancing Scalable and Reproducible Research
Building upon the data-centric foundation, CRADLE’s infrastructure also serves as a catalyst for scalable and reproducible research. It ensures thorough documentation of data and ML models to expand research efforts beyond the limitations imposed by dataset scale. The system eliminates the redundancy inherent in isolated studies by offering a platform that fosters the adoption of standardized data schemas, contributing to the development of models with greater generalizability.
A tangible demonstration of CRADLE’s impact is observed in the capabilities to analyze heterogeneous collections of XRD patterns, XCT scans, and AFM images. The five demonstrations were each built as an extension of a pre-existing work, where the previous study opted to only examine a subset of data due to volume constraints. Through CRADLE, we were able to extend analysis for the entire dataset to unlock new insights and workflows that scale. For instance, the computational efficiency achieved through the use of distributed computing processing 167 billion data points in about 4 h with Apache Spark and CRADLE’s Hadoop cluster highlights is illustrated in our benchmark analysis (see Fig. 4). This efficiency not only demonstrates CRADLE’s capacity to handle large-scale data analyses but also its role in facilitating more in-depth and expansive research in the field of materials science.
CRADLE’s adherence to FAIR principles in data transformation processes, coupled with the metadata storage alongside the transformed data, exemplifies the commitment to data and model provenance.[[48,49]] Leveraging open-source tooling, generating accessible repositories, and operating in collaborative scientific workflows highlight infrastructure’s role in advancing open science.
Conclusion
The paradigm shift towards data-centric AI is essential to fully utilize the potential of machine learning. Rather than refined algorithms, model efficacy fundamentally relies on quality and available data. CRADLE exemplifies this shift by prioritizing unified data management and accessibility first. This enables integrating fragmented stores and utilizing techniques like distributed computing and machine learning that otherwise fail with poor data systems. The focus moves beyond models to transform how data is captured, stored, and accessed across its lifecycle.
In this work, we demonstrate how the data-centric foundation provided by CRADLE enables analysis at new scales. Presented use cases highlighted techniques across modalities including large-scale inference for segmentation and characterization, accelerated deep learning with optimized data pipelines, multimodal integration across sensor streams, and distributed graph querying. From polymer crystallization studied with AFMs to satellite imagery monitoring, CRADLE catalyzes insights across domains. Future directions for CRADLE involve expanding its capabilities to support additional materials-specific software and making key middleware components portable and open source. CRADLE enables a data-centric AI paradigm that can help solve not only materials science challenges but other fields like biology, physics, and chemistry to push scientific discovery forward.
Data availability
Datasets that are open access are available at the OSF page of the CWRU SDLE Research Center.[[50]]
References
C. Draxl, M. Scheffler, NOMAD: the FAIR concept for big data-driven materials science. MRS Bull. 43, 676–682 (2018). https://doi.org/10.1557/mrs.2018.208
L. Himanen, A. Geurts, A.S. Foster, P. Rinke, Data-driven materials science: status, challenges, and perspectives. Adv. Sci. 6, 1900808 (2019). https://doi.org/10.1002/advs.201900808
The Minerals, Metals & Materials Society, Building a Materials Data Infrastructure: Opening New Pathways to Discovery and Innovation in Science and Engineering (The Minerals, Metals & Materials Society, 2017). https://doi.org/10.7449/mdistudy_1
R. Stevens, V. Taylor, J. Nichols, A.B. Maccabe, K. Yelick, D. Brown, AI for Science: Report on the Department of Energy (DOE) Town Halls on Artificial Intelligence (AI) for Science (Argonne National Lab (ANL), Argonne, 2020)
J. Carter, J. Feddema, D. Kothe, R. Neely, J. Pruet, R. Stevens, AI for Science, Energy, and Security Report (Argonne National Lab (ANL), Argonne, 2020)
M.H. Jarrahi, A. Memariani, S. Guha, The principles of data-centric AI. Commun. ACM 66, 84–92 (2023). https://doi.org/10.1145/3571724
T. Hope, D. Downey, D.S. Weld, O. Etzioni, E. Horvitz, A computational inflection for scientific discovery. Commun. ACM 66, 62–73 (2023). https://doi.org/10.1145/3576896
L. Aroyo, M. Lease, P.K. Paritosh, M. Schaekermann, Data excellence for AI: why should you care (2021), Preprint at https://arxiv.org/abs/2111.10391
A. Nihar, T. Ciardi, R. Chawla, O.D. Akanbi, V. Chaudhary, Y. Wu, R.H. French, Accelerating Time to Science Using CRADLE: A Framework for Materials Data Science (IEEE, Goa, 2023). https://doi.org/10.1109/HiPC58850.2023.00041
A. Khalilnejad, A.M. Karimi, S. Kamath, R. Haddadian, R.H. French, A.R. Abramson, Automated pipeline framework for processing of large-scale building energy time series data. PLoS ONE 15, 0240461 (2020). https://doi.org/10.1371/journal.pone.0240461
Y. Hu, V.Y. Gunapati, P. Zhao, D. Gordon, N.R. Wheeler, M.A. Hossain, T.J. Peshek, L.S. Bruckman, G. Zhang, R.H. French, A nonrelational data warehouse for the analysis of field and laboratory data from multiple heterogeneous photovoltaic test sites. IEEE J. Photovolt. 7(1), 230–236 (2017). https://doi.org/10.1109/JPHOTOV.2016.2626919
R. Arora, An Introduction to Big Data, High Performance Computing, High-Throughput Computing, and Hadoop, in Conquering Big Data with High Performance Computing. ed. by R. Arora (Springer International Publishing, Cham, 2016), pp.1–12. https://doi.org/10.1007/978-3-319-33742-5_1
K. Shvachko, H. Kuang, S. Radia, R. Chansler, The Hadoop Distributed File System, in 2010 IEEE 26th Symposium on Mass Storage Systems and Technologies MSST. (IEEE, 2010), pp.1–10. https://doi.org/10.1109/MSST.2010.5496972
J. Dean, S. Ghemawat, Mapreduce: Simplified Data Processing on Large Clusters, in OSDI’04: Sixth Symposium on Operating System Design and Implementation. (2004), pp. 137–150. https://doi.org/10.1145/1327452.132749
A. Auradkar, C. Botev, S. Das, D. De Maagd, A. Feinberg, P. Ganti, L. Gao, B. Ghosh, K. Gopalakrishna, B. Harris, J. Koshy, K. Krawez, J. Kreps, S. Lu, S. Nagaraj, N. Narkhede, S. Pachev, I. Perisic, L. Qiao, T. Quiggle, J. Rao, B. Schulman, A. Sebastian, O. Seeliger, A. Silberstein, Bb. Shkolnik, C. Soman, R. Sumbaly, K. Surlaker, S. Topiwala, C. Tran, B. Varadarajan, J. Westerman, Z. White, D. Zhang, J. Zhang, Data Infrastructure at Linkedin, in 2012 IEEE 28th International Conference on Data Engineering. (2012), pp. 1370–1381. https://doi.org/10.1109/ICDE.2012.147
Y. Fu, C. Soman, Real-Time Data Infrastructure at Uber, in Proceedings of the 2021 International Conference on Management of Data. (Association for Computing Machinery, New York, 2021), pp.2503–2516. https://doi.org/10.1145/3448016.3457552
K.R. Talley, R. White, N. Wunder, M. Eash, M. Schwarting, D. Evenson, J.D. Perkins, W. Tumas, K. Munch, C. Phillips, A. Zakutayev, Research data infrastructure for high-throughput experimental materials science. Patterns 2, 100373 (2021)
N. Brandt, L. Griem, C. Herrmann, E. Schoof, G. Tosato, Y. Zhao, P. Zschumme, M. Selzer, Kadi4Mat: a research data infrastructure for materials science. Data Sci. J. 20, 8 (2021). https://doi.org/10.5334/dsj-2021-008
J.E. Saal, S. Kirklin, M. Aykol, B. Meredig, C. Wolverton, Materials design and discovery with high-throughput density functional theory: the open quantum materials database (OQMD). JOM 65, 1501–1509 (2013). https://doi.org/10.1007/s11837-013-0755-4
S. Curtarolo, W. Setyawan, S. Wang, J. Xue, K. Yang, R.H. Taylor, L.J. Nelson, G.L.W. Hart, S. Sanvito, M. Buongiorno-Nardelli, N. Mingo, O. Levy, A distributed materials properties repository from high-throughput ab initio calculations. Comput. Mater. Sci. 58, 227–235 (2012). https://doi.org/10.1016/j.commatsci.2012.02.002
M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.W. Boiten, L.B. da Silva Santos, P.E. Bourne, J. Bouwman, A.J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C.T. Evelo, R. Finkers, A. Gonzalez-Beltran, A.J.G. Gray, P. Groth, C. Goble, J.S. Grethe, J. Heringa, P.A.C. Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S.J. Lusher, M.E. Martone, A. Mons, A.L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M.A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao, B. Mons, The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016)
B. Bayerlein, M. Schilling, H. Birkholz, M. Jung, J. Waitelonis, L. Mädler, H. Sack, PMD core ontology: achieving semantic interoperability in materials science. Mater. Design 237, 112603 (2024)
J. Blair, R.S. Canon, J. Deslippe, A. Essiari, A. Hexemer, A.A. MacDowell, , D.Y. Parkinson, S.J. Patton, L. Ramakrishnan, N. Tamura, B.L. Tierney, C.E. Tull, High Performance Data Management and Analysis for Tomography, in SPIE Optical Engineering + Applications, ed. by S.R. Stock (2014), p. 92121. https://doi.org/10.1117/12.2069862
L. Ramakrishnan, R.S. Canon, Experiences in building a data packaging pipeline for tomography beamline (2013).
A.V. Babu, T. Bicer, S. Kandel, T. Zhou, D.J. Ching, S. Henke, S. Veseli, R. Chard, A. Miceli, M.J. Cherukara, AI-assisted automated workflow for real-time X-ray ptychography data analysis via federated resources (2023), Preprint at https://doi.org/10.48550/arXiv.2304.0429
T. Bicer, D. Gursoy, R. Kettimuthu, I.T. Foster, B. Ren, V. De Andrede, F. De Carlo, Real-Time Data Analysis and Autonomous Steering of Synchrotron Light Source Experiments, in 2017 IEEE 13th International Conference on E-Science (e-Science). (IEEE, Auckland, 2017), pp.59–68. https://doi.org/10.1109/eScience.2017.53
V.K. Vavilapalli, A.C. Murthy, C. Douglas, S. Agarwal, M. Konar, R. Evans, T. Graves, J. Lowe, H. Shah, S. Seth, B. Saha, C. Curino, O. O’Malley, S. Radia, B. Reed, Apache Hadoop Yarn: Yet Another Resource Negotiator, in Proceedings of the 4th Annual Symposium on Cloud Computing. (Association for Computing Machinery, New York, 2013), pp.1–16. https://doi.org/10.1145/2523616.2523633
A.B. Yoo, M.A. Jette, M. Grondona, Slurm: Simple Linux Utility for Resource Management, in Job Scheduling Strategies for Parallel Processing. ed. by D. Feitelson, L. Rudolph, U. Schwiegelshohn (Springer, Berlin, Heidelberg, 2003), pp.44–60. https://doi.org/10.1007/10968987_3
M. Zaharia, R.S. Xin, P. Wendell, T. Das, M. Armbrust, A. Dave, X. Meng, J. Rosen, S. Venkataraman, M.J. Franklin, A. Ghodsi, J. Gonzalez, S. Shenker, I. Stoica, Apache spark: a unified engine for big data processing. Commun. ACM 59, 56–65 (2016). https://doi.org/10.1145/2934664
M. Zaharia, M. Chowdhury, T. Das, A. Dave, J. Ma, M. McCauley, M.J. Franklin, S. Shenker, I. Stoica, Resilient Distributed Datasets: A Fault-tolerant Abstraction for In-memory Cluster Computing, in Proceedings of the 9th USENIX Conference on Networked Systems Design and Implementation. NSDI’12, (USENIX Association, USA, 2012), p. 2. https://doi.org/10.5555/2228298.222830
J.E. Gonzalez, R.S. Xin, A. Dave, D. Crankshaw, M.J. Franklin, I. Stoica, Graphx: Graph Processing in a Distributed Dataflow Framework, in Proceedings of the 11th USENIX Conference on Operating Systems Design and Implementation. OSDI’14, (USENIX Association, USA, 2014), pp. 599–613. https://doi.org/10.5555/2685048.268509
M. Kornacker, A. Behm, V. Bittorf, T. Bobrovytsky, C. Ching, A. Choi, J. Erickson, M. Grund, D. Hecht, M. Jacobs, I. Joshi, L. Kuff, D. Kumar, A. Leblang, N. Li, I. Pandis, H. Robinson, D. Rorke, S. Rus, J. Russell, D. Tsirogiannis, S. Wanderman-Milne, M. Yoder, Impala: A Modern, Open-source Sql Engine for Hadoop, in Conference on Innovative Data Systems Research, (2015)
J.G. Steiner, B.C. Neuman, J. Schiller, Kerberos: an authentication service for open network systems (1988). https://www.semanticscholar.org/paper/Kerberos%3A-An-Authentication-Service-for-Open-Steiner-Neuman/2c4aff896cd8e60b1ad59c02952947700ebc8edf. Accessed 18 Jul 2023
G.M. Kurtzer, V. Sochat, M.W. Bauer, Singularity: scientific containers for mobility of compute. PLOS ONE 12(5), 1–20 (2017). https://doi.org/10.1371/journal.pone.0177459
M. Abadi, P. Barham, J. Chen, Z. Chen, A. Davis, J. Dean, M. Devin, S. Ghemawat, G. Irving, M. Isard, M. Kudlur, J. Levenberg, R. Monga, S. Moore, D.G. Murray, B. Steiner, P. Tucker, V. Vasudevan, P. Warden, M. Wicke, Y. Yu, X. Zheng, Tensorflow: A System for Large-Scale Machine Learning. in Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation 16, (USENIX Association, Savannah, 2016), pp. 265–283
A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Köpf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai, S. Chintala, PyTorch: an imperative style, high-performance deep learning library (2019), Preprint at https://arxiv.org/abs/1912.01703
D. Hudak, D. Johnson, A. Chalker, J. Nicklas, E. Franz, T. Dockendorf, B. McMichael, Open ondemand: a web-based client portal for HPC centers. J. Open Source Softw. 3, 622 (2018). https://doi.org/10.21105/joss.00622
W. Yue, P.K. Tripathi, G. Ponon, Z. Ualikhankyzy, D.W. Brown, B. Clausen, M. Strantza, D.C. Pagan, M.A. Willard, F. Ernst, E. Ayday, V. Chaudhary, R.H. French, Phase identification in synchrotron X-ray diffraction patterns of Ti–6Al–4V using computer vision and deep learning. Integr. Mater. Manuf. Innov. (2024). https://doi.org/10.1007/s40192-023-00328-0
W. Yue, M.R. Mehdi, P.K. Tripathi, M.A. Willard, F. Ernst, R.H. French, Exploring 2D X-ray diffraction phase fraction analysis with convolutional neural networks: Insights from kinematic-diffraction simulations. MRS Adv. (2024). https://doi.org/10.1557/s43580-024-00862-9
M.S. Kalutotage, T.G. Ciardi, P.K. Tripathi, L. Huang, J.C. Jimenez, P.J. Noell, L.S. Bruckman, R.H. French, A. Sehirligolu, Automated Image Segmentation and Processing Pipeline Applied to X-ray Computed Tomography Studies of Pitting Corrosion in Aluminum Wires. Under review
O. Ronneberger, P. Fischer, T. Brox, U-net: Convolutional Networks for Biomedical Image Segmentation, in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015. ed. by N. Navab, J. Hornegger, W.M. Wells, A.F. Frangi (Springer International Publishing, Cham, 2015), pp.234–241. https://doi.org/10.1007/978-3-319-24574-4_28
M. Lu, S.N. Venkat, J. Augustino, D. Meshnick, J.C. Jimenez, P.K. Tripathi, A. Nihar, C.A. Orme, R.H. French, L.S. Bruckman, Y. Wu, Image processing pipeline for fluoroelastomer crystallite detection in atomic force microscopy images. Integr. Mater. Manuf. Innov. (2023). https://doi.org/10.1007/s40192-023-00320-8
S.N. Venkat, T. Ciardi, M. Lu, J. Augustino, A. Goodman, P. DeLeo, P.K. Tripathi, J.C. Jimenez, A. Mondal, F. Ernst, C.A. Orme, Y. Wu, R.H. French, L.S. Bruckman, A general materials data science framework for quantitative 2D analysis of crystallization kinetics of particle growth from image sequences. Mater. Manuf. Innov. Integr. (2024). https://doi.org/10.1007/s40192-024-00342-w
K. Hernandez, T. Ciardi, R. Yamamoto, M. Lu, A. Nihar, J. Jimenez, P. Tripathi, B. Giera, J.B. Forien, J. Lewandowski, R. French, L. Bruckman, L-PBF high-throughput data pipeline approach for multi-modal integration. Integr. Mater. Manuf. Innov. (2024). https://doi.org/10.1007/s40192-024-00368-0
K.J. Hernandez, E.I. Barcelos, J.C. Jimenez, A. Nihar, P.K. Tripathi, B. Giera, R.H. French, L.S. Bruckman, A data integration framework of additive manufacturing based on FAIR principles. MRS Adv. (2024). https://doi.org/10.1557/s43580-024-00874-5
M. Ning, Y. Lu, W. Hou, M. Matskin, Yolov4-Object: An Efficient Model and Method for Object Discovery, in 2021 IEEE 45th Annual Computers, Software, and Applications Conference (COMPSAC). (IEEE, 2021), pp.31–36. https://doi.org/10.1109/COMPSAC51774.2021.00016
O.D. Akanbi, D.C. Bhuvanagiri, E.I. Barcelos, A. Nihar, B. Gonzalez Hernandez, J.M. Yarus, R.H. French, Integrating multiscale geospatial analysis for monitoring crop growth, nutrient distribution, and hydrological dynamics in large-scale agricultural systems. J. Geovisualization Spatial Anal. 8, 9 (2024). https://doi.org/10.1007/s41651-023-00164-y
M. Lu, L. Huang, W.C. Oltjen, X. Yu, A Nihar, T.G. Ciardi, E. Barcelos, P. Tripathi, A. Daundkar, D. Bhuvanagiri, H. Omodolor, O. Akanbi, H.H. Aung, K.J. Hernandez, M.M. Rasmussen, R.J. Wieser, S.N. Venkat, T. Wang, W. Yue, Y. Fan, R. Chawla, L. Jo, Z. Li, J. Liu, J.P. Glynn, K.A. Coleman, J.M. Yarus, M. Li, K.O. Davis, L.S. Bruckman, Y. Wu, R.H. French, FAIRmaterials: generate Json-Ld format files based on FAIRification standard (2023), https://pypi.org/project/fairmaterials/. Accessed 30 Dec 2022
P. Rajamohan, A.H. Bradley, H. Caldwell, E.I. Barcelos, R.H. French, FAIRmaterials: Find the Docs (2023), https://cwrusdle.bitbucket.io/. Accessed 14 Mar 2023
R.H. French, A. Nihar, E. Barcelos, R. Wieser, A. Curran, A.M Karimi, J.L. Braid, D. Gordon, J. Liu, M. Wang, CWRU SDLE Research Center. (OSF, 2019), https://osf.io/wn35j/. Accessed 09 Mar 2023
Funding
This work was supported in part by NSF award 2117439 and DOE NNSA award DE-NA0004104. It also used the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University.
Author information
Authors and Affiliations
Contributions
The authors confirm contribution to the paper as follows: study conception, design: AN, RF, VC, PT, YW; software specification, design, development, and deployment: AN, TGC, RC; draft manuscript preparation: TGC, RC; use case implementation: TGC, OA. All authors reviewed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ciardi, T.G., Nihar, A., Chawla, R. et al. Materials data science using CRADLE: A distributed, data-centric approach. MRS Communications 14, 601–611 (2024). https://doi.org/10.1557/s43579-024-00616-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1557/s43579-024-00616-6