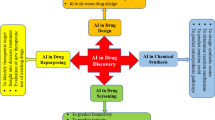
Abstract
Over the last decade, deep learning (DL) methods have been extremely successful and widely used to develop artificial intelligence (AI) in almost every domain, especially after it achieved its proud record on computational Go. Compared to traditional machine learning (ML) algorithms, DL methods still have a long way to go to achieve recognition in small molecular drug discovery and development. And there is still lots of work to do for the popularization and application of DL for research purpose, e.g., for small molecule drug research and development. In this review, we mainly discussed several most powerful and mainstream architectures, including the convolutional neural network (CNN), recurrent neural network (RNN), and deep auto-encoder networks (DAENs), for supervised learning and nonsupervised learning; summarized most of the representative applications in small molecule drug design; and briefly introduced how DL methods were used in those applications. The discussion for the pros and cons of DL methods as well as the main challenges we need to tackle were also emphasized.





Similar content being viewed by others
Change history
25 June 2018
The name of the corresponding author should be ‘Xiang-Qun Xie’, rather than ‘Xiang-Qun Sean Xie’.
References
Artificial intelligence: Google’s AlphaGo beats Go master Lee Se-dol. In: Technology. BBC NEWS. 12 March 2016. http://www.bbc.com/news/technology-35785875#. Accessed 15 Dec 2017.
Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, et al. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529(7587):484–9.
Ma C, Wang L, Xie X-Q. GPU accelerated chemical similarity calculation for compound library comparison. J Chem Inf Model. 2011;51(7):1521–7.
Baskin II, Winkler D, Tetko IV. A renaissance of neural networks in drug discovery. Expert Opin Drug Discov. 2016;11(8):785–95.
McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5(4):115–33.
Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323:533–6.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44.
Gao B, Xu Y. Univariant approximation by superpositions of a sigmoidal function. J Math Anal Appl. 1993;178(1):221–6.
Lawrence S, Giles CL. Overfitting and neural networks: conjugate gradient and backpropagation. In: Neural Networks, 2000. IJCNN 2000, Proceedings of the IEEE-INNS-ENNS International Joint Conference, Como, Italy. 2000. Vol. 1, pp. 114–19.
Hochreiter S. The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int J Uncertainty Fuzziness Knowledge Based Syst. 1998;6(2):107–16.
Winkler DA, Le TC. Performance of deep and shallow neural networks, the universal approximation theorem, activity cliffs, and QSAR. Mol Inf. 2017;36(1–2):1600118.
Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18:1527–54.
Olurotimi O. Recurrent neural network training with feedforward complexity. IEEE Trans Neural Netw. 1994;5(2):185–97.
Cox DR. The regression-analysis of binary sequences. J R Stat Soc Ser B Stat Methodol. 1958;20(2):215–42.
Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–97.
Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn. 1997;29(2–3):103–30.
Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–58.
Goh GB, Hodas NO, Vishnu A. Deep learning for computational chemistry. J Comput Chem. 2017;38(16):1291–307.
Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol Inf. 2010;29(6–7):476–88.
Chen B, Sheridan RP, Hornak V, Voigt JH. Comparison of random forest and Pipeline Pilot naïve Bayes in prospective QSAR predictions. J Chem Inf Model. 2012;52:792–803.
Myint KZ, Xie X-Q. Ligand biological activity predictions using fingerprint-based artificial neural networks (FANN-QSAR). Methods Mol Biol (Clifton, NJ). 2015;1260:149–64.
Ma C, Wang L, Yang P, Myint KZ, Xie XQ. LiCABEDS II. Modeling of ligand selectivity for G-protein coupled cannabinoid receptors. J Chem Inf Model. 2013;53(1):11–26.
Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43(D1):D1079.
Alexander DL, Tropsha A, Winkler DA. Beware of R(2): simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J Chem Inf Model. 2015;55(7):1316–22.
Bengio Y. Learning deep architectures for AI. Found Trends® Mach Learn. 2009;2(1):1–127.
Ekins S. The next era: deep learning in pharmaceutical research. Pharm Res. 2016;33(11):2594–603.
Gawehn E, Hiss JA, Schneider G. Deep learning in drug discovery. Mol Inf. 2016;35(1):3–14.
Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69(Supplement):S36–40.
Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol Pharm. 2016;13(5):1445–54.
Pastur-Romay AL, et al. Deep artificial neural networks and neuromorphic chips for big data analysis: pharmaceutical and bioinformatics applications. Int J Mol Sci. 2016;17(8):E1313.
van Westen GJP, Wegner JK, IJzerman AP, van Vlijmen HWT, Bender A. Proteochemometric modeling as a tool to design selective compounds and for extrapolating to novel targets. Med Chem Comm. 2011;2(1):16–30.
Rosenblatt F. The perceptron, a perceiving and recognizing automaton project para. Buffalo: Cornell Aeronautical Laboratory; 1957. Vol. 85, pp. 460–61.
Kelley HJ. Gradient theory of optimal flight paths. Ars J. 1960;30(10):947–54.
Google supercharges machine learning tasks with TPU custom chip. 2016 [cited 2017 May 20th].
Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117.
Hubel DH, Wiesel TN. Receptive fields, binocular interaction and functional architecture in the cat's visual cortex. J Physiol. 1962;160(1):106–54.
Hubel DH, Wiesel TN. Receptive fields of single neurones in the cat’s striate cortex. J Physiol. 1959;148(3):574–91.
Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. In: European conference on computer vision 2014 Sep 6. Cham: Springer; 2014. pp. 818–33.
Lecun Y, Jackel LD, Bottou L, Brunot A, Cortes C, Denker JS, et al. Comparison of learning algorithms for handwritten digit recognition. In: Fogelman F, Gallinari P, editors. International conference on artificial neural networks. Paris: EC2 & Cie. 1995. p. 53–60.
LeCun Y, et al. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–324.
Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, Bengio Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. In: Conference on empirical methods in natural language processing, Doha, Qatar. 2014. Vol. 1, pp. 1724–34.
Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–80.
Si S. Hsieh C Dhillon I, Proceedings of the 31st international conference on machine learning. 2014.
Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7.
Chen Y, Lin Z, Zhao X, Wang G, Gu Y. Deep learning-based classification of hyperspectral data. IEEE J Sel Topics Appl Earth Observ Remote Sens. 2014;7(6):2094–107.
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Advances in neural information processing systems. 2014. pp. 2672–80.
Srivastava N, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929.
Burden F, Winkler D. Bayesian regularization of neural networks. Methods Mol Biol. 2008;458:25–44.
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–52.
Unterthiner T, Mayr A, Klambauer G, Steijaert M, Wegner JK, Ceulemans H, Hochreiter S. Deep learning as an opportunity in virtual screening. In: Proceedings of the deep learning workshop at NIPS, 2014 Dec 8. Vol. 27, pp. 1-9.
Casey W. Tox21 overview and update. In Vitro Cell Dev Biol Anim. 2013;49:S7–8.
Wu Z, Ramsundar B, Feinberg EN, Gomes J, Geniesse C, Pappu AS, Leswing K, Pande V. MoleculeNet: a benchmark for molecular machine learning. Chem Sci. 2018;9(2):513–30.
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(D1):D1100–7.
Lenselink EB, ten Dijke N, Bongers B, Papadatos G, van Vlijmen HWT, Kowalczyk W, et al. Beyond the hype: deep neural networks outperform established methods using a ChEMBL bioactivity benchmark set. J Cheminform. 2017;9(1):45.
Rubio DM, Schoenbaum EE, Lee LS, Schteingart DE, Marantz PR, Anderson KE, et al. Defining translational research: implications for training. Acad Med: J Assoc Am Med Coll. 2010;85(3):470–5.
Wang Y, Zeng J. Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics. 2013;29(13):i126–34.
Ma J, Sheridan RP, Liaw A, Dahl GE, Svetnik V. Deep neural nets as a method for quantitative structure–activity relationships. J Chem Inf Model. 2015;55(2):263–74.
Dahl GE, Jaitly N, Salakhutdinov R. Multi-task neural networks for QSAR predictions. arXiv preprint in Machine Learning (stat.ML). arXiv:1406.1231. 2014 Jun 4.
Ramsundar B, Kearnes S, Riley P, Webster D, Konerding D, Pande V. Massively multitask networks for drug discovery. arXiv preprint in Machine Learning (stat.ML). arXiv:1502.02072. 2015 Feb 6.
Wang C, Liu J, Luo F, Tan Y, Deng Z, Hu QN. Pairwise input neural network for target-ligand interaction prediction. In: 2014 I.E. International Conference on Bioinformatics and Biomedicine (BIBM), 2014 Nov 2. IEEE. pp. 67–70.
Wallach I, Dzamba M, Heifets A. Atomnet: A deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv preprint in Learning (cs.LG). arXiv:1510.02855. 2015 Oct 10.
Wan F, Zeng J. Deep learning with feature embedding for compound-protein interaction prediction. bioRxiv. 2016. https://doi.org/10.1101/086033.
Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50(5):742–54.
Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. In: Proceedings of the 27th international conference on machine learning (ICML-10), 2010. pp. 807–14.
Lusci A, Pollastri G, Baldi P. Deep architectures and deep learning in chemoinformatics: the prediction of aqueous solubility for drug-like molecules. J Chem Inf Model. 2013;53(7):1563–75.
Shin M, Jang D, Nam H, Lee KH, Lee D. Predicting the absorption potential of chemical compounds through a deep learning approach. IEEE/ACM Trans Comput Biol Bioinform. 2016;PP(99):1–1.
Mayr A, Klambauer G, Unterthiner T, Hochreiter S. DeepTox: Toxicity Prediction using Deep Learning. Front Environ Sci. 2016;3(80). https://doi.org/10.3389/fenvs.2015.00080.
Pereira JC, Caffarena ER, Dos Santos CN. Boosting docking-based virtual screening with deep learning. J Chem Inf Model. 2016;56(12):2495–506.
Hinton GE, McClelland JL, Rumelhart DE. Distributed representations. In: Parallel distributed processing: explorations in the microstructure of cognition. Cambridge: MIT Press; 1986. Vol. 1, No. 3, pp. 77–109.
Yao K, Parkhill J. Kinetic energy of hydrocarbons as a function of electron density and convolutional neural networks. J Chem Theory Comput. 2016;12(3):1139–47.
Bjerrum EJ. Smiles enumeration as data augmentation for neural network modeling of molecules. arXiv preprint in Learning (cs.LG). arXiv:1703.07076. 2017 Mar 21.
Weininger D. Smiles, a chemical language and information-system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28(1):31–6.
Goh GB, Siegel C, Vishnu A, Hodas NO, Baker N. Chemception: A deep neural network with minimal chemistry knowledge matches the performance of expert-developed qsar/qspr models. arXiv preprint in Machine Learning (stat.ML). arXiv:1706.06689. 2017 Jun 20.
Goh GB, Siegel C, Vishnu A, Hodas NO, Baker N. How much chemistry does a deep neural network need to know to make accurate predictions? arXiv preprint in Machine Learning (stat.ML). arXiv:1710.02238. 2017 Oct 5.
Kadurin A, Aliper A, Kazennov A, Mamoshina P, Vanhaelen Q, Khrabrov K, et al. The cornucopia of meaningful leads: applying deep adversarial autoencoders for new molecule development in oncology. Oncotarget. 2017;8(7):10883–90.
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(Database issue):D1202–13.
Segler MHS, Kogej T, Tyrchan C, Waller MP. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Central Science. 2018;4(1):120–31.
Olivecrona M, Blaschke T, Engkvist O, Chen H. Molecular de-novo design through deep reinforcement learning. Journal of Cheminformatics. 2017;9(1):48.
Lima Guimaraes G, Sanchez-Lengeling B, Cunha Farias PL, Aspuru-Guzik A. Objective-reinforced generative adversarial networks (ORGAN) for sequence generation models. arXiv preprint in Machine Learning (stat.ML). arXiv:1705.10843. 2017 May.
Capuzzi SJ, et al. QSAR modeling of Tox21 challenge stress response and nuclear receptor signaling toxicity assays. Front Environ Sci. 2016;4(3):45.
Maggiora GM. On outliers and activity cliffs—why QSAR often disappoints. J Chem Inf Model. 2006;46(4):1535.
Myint K-Z, Wang L, Tong Q, Xie XQ. Molecular fingerprint-based artificial neural networks QSAR for ligand biological activity predictions. Mol Pharm. 2012;9(10):2912–23.
Wang L, Ma C, Wipf P, Liu H, Su W, Xie XQ. TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database. AAPS J. 2013;15(2):395–406.
Kearnes S, McCloskey K, Berndl M, Pande V, Riley P. Molecular graph convolutions: moving beyond fingerprints. J Comput Aided Mol Des. 2016;30(8):595–608.
Hughes TB, Dang NL, Miller GP, Swamidass SJ. Modeling reactivity to biological macromolecules with a deep multitask network. ACS Cent Sci. 2016;2(8):529–37.
Hughes TB, Miller GP, Swamidass SJ. Modeling epoxidation of drug-like molecules with a deep machine learning network. ACS Cent Sci. 2015;1(4):168–80.
Acknowledgements
The authors thank Dr. Yuanqiang Wang, Nan Wu, and Yubin Ge in the CCGS Center at the University of Pittsburgh (Pitt) for carefully reviewing the manuscripts and providing helpful comments for revision. Thanks to all the students and faculty in the CDAR Center, School of Pharmacy at Pitt for their help and supports. The authors also acknowledge the funding support to our laboratory from NIH NIDA (P30DA035778) and DOD (W81XWH-16-1-0490).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jing, Y., Bian, Y., Hu, Z. et al. Deep Learning for Drug Design: an Artificial Intelligence Paradigm for Drug Discovery in the Big Data Era. AAPS J 20, 58 (2018). https://doi.org/10.1208/s12248-018-0210-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-018-0210-0