Abstract
Background
Carcinogens in cigarette smoke may cause aberrant epigenomic changes. The hypomethylation of long interspersed nucleotide element-1 (LINE-1) in colorectal carcinoma has been associated with genomic instability and worse clinical outcome. We hypothesized that the association between smoking behavior and colorectal cancer mortality might be stronger in tumors with lower LINE-1 methylation levels.
Findings
To test our hypothesis, we examined the interaction of tumor LINE-1 methylation levels and smoking status at diagnosis using data of 1208 cases among 4420 incident colorectal cancer cases that were ascertained in two prospective cohort studies. We conducted multivariable Cox proportional hazards regression analyses, using inverse probability weighting with covariate data of the 4420 cases to control for potential confounders and selection bias due to data availability. The prognostic association of smoking status at diagnosis differed by tumor LINE-1 methylation levels (Pinteraction = 0.050 for overall mortality and 0.017 for colorectal cancer-specific mortality; with an alpha level of 0.005). In cases with <60% LINE-1 methylation, current smoking (vs. never smoking) was associated with worse overall mortality (multivariable hazard ratio, 1.80; 95% confidence interval, 1.19–2.73). In contrast, smoking status was not associated with mortality in cases with ≥60% LINE-1 methylation.
Conclusions
Our findings suggest that the association between smoking status and mortality is stronger in colorectal cancer patients with lower tumor LINE-1 methylation levels. These results warrant further investigation into an interactive role of smoking and aberrant DNA methylation in colorectal cancer progression.
Similar content being viewed by others
Introduction
Cigarette smoke is an established risk factor for colorectal cancer incidence and mortality [1,2,3]. Cigarette smoke contains hundreds of carcinogens, some of which can cause epigenetic alterations [4]. While there is strong evidence linking smoking to epigenetic changes, including findings from Epigenome-Wide Association Studies, less is known about epigenetic changes in former smokers, particularly long-time quitters [5, 6].
Accumulating evidence suggests that DNA hypomethylation may play an important role in colorectal cancer progression [7, 8]. Long interspersed nucleotide element-1 (LINE-1) hypomethylation is a surrogate marker for genome-wide DNA hypomethylation [9], and LINE-1 hypomethylated colorectal cancer has been associated with worse prognosis and non-response to certain chemotherapies, suggesting potential utilization of LINE-1 hypomethylation as a prognostic biomarker [10, 11]. Moreover, LINE-1 hypomethylation is an emerging biomarker for early-onset colorectal cancer diagnosed before age 50 [12], which has shown increasing incidence in many parts of the world since the 1980s [13].
To test whether the association of smoking status at diagnosis with colorectal cancer mortality might differ by LINE-1 methylation levels in tumors, we leveraged data from two large prospective cohorts with 4420 incident colorectal cancer cases, including 1208 cases with available tumor tissue data.
Methods
Study population and design
The Nurses’ Health Study (NHS, N = 121,701) was established in 1976 and the Health Professionals Follow-up Study (HPFS, N = 51,529) was established in 1986 [14, 15]. Self-administered questionnaires were mailed to participants at baseline and then biennially to update smoking status, lifestyle, and medical history. Semi-quantitative food frequency questionnaires were administered every 4 years to assess participants’ diet.
Deaths were identified through next-of-kin reports or the National Death Index, and cause of death was determined by study physicians after a review of the medical records or death certificates. A pathologist (S.O.), blinded to other information, conducted a centralized review of hematoxylin and eosin (H&E)-stained tissue sections of all colorectal cancer cases and recorded pathological features including tumor differentiation. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and the Harvard T. H. Chan School of Public Health, and those of participating registries as required. We also obtained signed consents from patients (or next-of-kin for deceased patients) to use CRC tissue specimens for molecular pathological analyses.
Assessment of smoking behavior at diagnosis
Detailed information on smoking was obtained as previously described [1, 2, 15]. Smoking status was divided into three categories (never smoking, past smoking, and current smoking at the time of diagnosis). As most current smokers (86%) had a smoking history of ≥20 packyears at diagnosis, we did not examine associations by packyears of smoking in current smokers. Of the 599 past smokers, 285 (47.6%) had a smoking history of 1–19 packyears and 314 cases (52.4%) had a history of packyears of ≥20. Past smokers who quit ≥10 years prior to diagnosis were more likely to have a smoking history of 1–19 packyears than those who quit <10 years prior to diagnosis (201 out of 322 or 62% vs. 84 out of 277 or 30%); therefore, for past smokers, we did not further stratify by time since quitting smoking (Additional file 1: Supplementary Table 1). Smoking status, diet, and lifestyle at the time of diagnosis were defined using participants’ most recent available questionnaire prior to the diagnosis of cancer.
Analyses of LINE-1 methylation analysis, microsatellite instability (MSI), CpG island methylator phenotype (CIMP), and KRAS, BRAF, and PIK3CA mutations
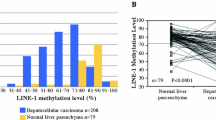
More details on our methods and pyrosequencing results were described in our previous publications [11, 16]. In brief, DNA was extracted from formalin-fixed paraffin-embedded tissue blocks, focusing on tumor areas only. We performed bisulfite DNA treatment, polymerase chain reaction (PCR), and pyrosequencing using the PyroMark kit (Qiagen), and quantified LINE-1 methylation levels by amplifying a region of the LINE-1 element (position 305 to 331 in accession No. X58075) which includes 4 CpG sites. We used the average of the proportions of C nucleotides at the 4 CpG sites (a scale of 0 to 100) as the LINE-1 methylation levels of each case. The LINE-1 methylation level showed a normal distribution [16] and was used both as a continuous variable (scale 0–100%) and a categorical variable [“high” (≥68% methylation), “intermediate” (≥60% and <68% methylation), and “low” (<60% methylation)]. The preciseness of pyrosequencing assay has been validated in our previous study using approximately 500 cancer cells from 5 anonymized colorectal cancer cases which were collected by laser capture microdissection [17].
MSI analysis was carried out utilizing a panel of 10 microsatellite markers, as previously described [18]. MSI-high was defined as instability in ≥30% of the markers. We quantified DNA methylation in eight CpG island methylator phenotype (CIMP)-specific promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) by using bisulfite DNA treatment and real-time PCR (MethyLight) as previously described [19, 20]. PCR and pyrosequencing targeted for KRAS (codons 12, 13, 61, and 146) [21, 22], BRAF (codon 600) [18], and PIK3CA (exons 9 and 20) were performed as previously described [23].
Statistical analyses
Our primary hypothesis testing was an assessment of a statistical interaction between smoking status at diagnosis [ordinal categories; never smoker (0), past smoker with 1–19 packyears (1), past smoker with ≥ 20 packyears (2), and current smoker (3)] and tumor LINE-1 methylation levels (continuous) using Cox proportional hazards regression. We utilized the Wald test to assess the statistical significance of that interaction and reported the P value as Pinteraction. Our main endpoints were all-cause and colorectal cancer-specific mortality. Survival time was defined as the time from colorectal cancer diagnosis until death or the end of follow-up, whichever came first (January 1, 2016, for the HPFS; May 31, 2016, for the NHS). For the analysis of colorectal cancer-specific mortality, deaths from other causes were censored. We calculated hazard ratio (HR) and its 95% confidence interval (CI) using re-parameterization of the interaction term in a single regression model. The trend test was conducted using the ordinal smoking variable.
To reduce potential selection bias due to tumor tissue data availability, we applied the inverse probability weighting (IPW) method using all 4420 cases as reported previously [24, 25].
We used the multivariable IPW-adjusted Cox proportional hazards regression models to adjust for potential confounders, which initially included age at diagnosis, year of diagnosis of cancer family history of colorectal cancer in first degree relatives, body mass index at diagnosis (BMI), alcohol consumption at diagnosis, empirical dietary inflammatory pattern (EDIP) score at diagnosis, dietary fiber intake at diagnosis, folate intake at diagnosis, regular aspirin use at diagnosis, physical activity at diagnosis, tumor location and differentiation, MSI and CIMP status, and KRAS, BRAF, and PIK3CA mutations. We conducted a backward elimination with a threshold of P = 0.05 to select variables for the final models. The proportionality of hazards assumption was generally satisfied after assessment of Schoenfeld residual plots and including interaction terms of smoking and survival time to the multivariable models. All P values were two-sided and a P <0.005 was considered statistically significant as recommended by the expert panel [26].
Results
Patients’ characteristics
With a follow-up to 2012, we documented 4420 incident colorectal cancer cases including 1208 patients with available data on both smoking information at diagnosis and tumor LINE-1 methylation level (Additional file 1: Supplementary Table 2). After a median follow-up time of 16 years (interquartile range 11.9–20.3 years) for censored cases, 776 all-cause deaths including 343 colorectal cancer-specific deaths were identified.
Smoking status and mortality
Compared with never smokers, HRs of past smokers with <19 packyears, past smokers with ≥20 packyears, and current smokers were 0.81 (0.65–1.00), 1.01 (0.85–1.21), and 1.31 (0.99–1.71) for all-cause mortality and 0.79 (0.58–1.08), 0.86 (0.65–1.13), and 1.07 (0.75–1.53) for colorectal cancer-specific mortality, respectively (Additional file 1: Supplementary Table 3).
Tumor LINE-1 methylation levels and mortality
Colorectal cancer cases who developed tumors with high LINE-1 methylation level tumors (≥68%) appeared to have a lower risk of colorectal cancer-specific mortality (HR 0.74, 95% CI 0.55–0.99) than those with low (<60%) LINE-1 methylation levels. However, the lower risk in cases with high LINE-1 methylation level tumors was not seen for all-cause mortality (Additional file 1: Supplementary Table 4).
Smoking and mortality in relation to tumor LINE-1 methylation levels
In our primary hypothesis testing, the association between smoking status and mortality appeared to be stronger in cases with low levels of LINE-1 methylation tumors than in those with high levels of LINE-1 methylation (Pinteraction = 0.050 for all-cause mortality, Pinteraction = 0.017 for colorectal cancer-specific mortality; Table 1). Among cases with low LINE-1 methylation tumors (< 60%), HRs of current smoking relative to never smoking were 1.55 (95% CI, 0.94–2.56) for colorectal cancer mortality and 1.80 (95% CI, 1.19–2.73) for all-cause mortality. On the other hand, among cases with high levels of LINE-1 methylation (≥68%), corresponding HRs (current vs. never smoking) were 0.93 (95% CI, 0.50–1.73) for colorectal cancer-specific mortality and 1.33 (95% CI, 0.85–2.08) for all-cause mortality. Results did not change substantially when we repeated analyses without using IPW adjustment, or restricted analyses to cases with stage I–III colorectal cancers (Additional files 1: Supplementary Tables 5 and 6).
Discussion
Previous studies have suggested that smoking may be associated with worse survival in colorectal cancer, perhaps in certain tumor molecular subtypes though findings have not been consistent [27,28,29,30]. In contrast to those tumor markers (except for MSI status) in the previous studies [27,28,29], tumor LINE-1 hypomethylation has been consistently shown to be a strong prognostic indicator in various cancer types including colorectal cancer [12, 31, 32]. Our findings suggest that the positive association between smoking status at diagnosis and mortality may be more pronounced in cases with low LINE-1 methylation levels than in those with high LINE-1 methylation levels.
Experimental studies have shown that DNMT1 (DNA methyltransferase 1) mutation can lead to global DNA hypomethylation [7] and that global DNA hypomethylation promotes tumor development through chromosomal instability including loss of heterozygosity of TP53, which can cause cell cycle arrest for DNA repair or apoptosis of damaged cells [33]. Assuming that tumors with high genomic instability are more prone to somatic mutation induced by exogenous factors such as smoking, it is possible that cases who later on developed tumors with LINE-1 hypomethylation may have been more susceptible to the mutagenic effects of smoking than those with high LINE-1 methylation tumors. Our findings suggest that the stronger association between smoking and mortality observed in LINE-1 hypomethylated tumors may be, at least in part, explained by a higher accumulation of genomic instability over time. In addition, tumor LINE-1 hypomethylation has been associated with lower levels of T cell immune response to colorectal cancer, suggesting its immunosuppressive effect [34]. Furthermore, smoking has been associated with the incidence of colorectal cancer subtypes containing fewer counts of T cells and macrophages, implying its suppressive effect on effector immune cells [35, 36].
Together with these previous findings, our current results may suggest that smoking status and tumor LINE-1 hypomethylation interact and jointly influence the tumor-immune interaction, leading to a stronger prognostic role of smoking status for tumors with LINE-1 hypomethylation.
Strength and limitations
One major strength of this study was our molecular pathological epidemiology [37,38,39] database of colorectal cancer cases with the availability of diet and lifestyle information that has been prospectively and repeatedly collected. This rich database enabled us to examine the prognostic interaction between smoking behavior and tumor LINE-1 methylation levels while adjusting for multiple potential confounders [40, 41] and selection bias due to tumor molecular data availability [25].
There are several limitations to our study. First, we did not examine the effect of postdiagnosis smoking status because 40% of current smokers (44 out of 110) quit smoking after diagnosis and the sample size of postdiagnosis current smokers was small. Second, data on cancer treatment were limited in this dataset. However, it is unlikely that the ratio of patients who underwent chemotherapy differed substantially according to smoking status. Additionally, treatment protocols for colorectal cancers are generally similar across the USA and adjusting for the AJCC stage should have limited potential confounding due to treatment. Third, data on cancer recurrence were unavailable. However, given a follow-up of >10 years, colorectal cancer-specific mortality can be considered as a reasonable measure of colorectal cancer outcome. Fourth, our main results did not meet our stringent multiple comparison-adjusted significance level of P < 0.005. However, we selected all the risk factors and statistical comparisons on the basis of previous data and certain hypotheses and interpreted our results prioritizing biological plausibility, coherence, and consistency rather than only statistical significance. Fifth, there is evidence that LINE-1 hypomethylation is inversely associated with MSI-high, CIMP-high, and BRAF-mutated CRC [11, 16]. While we adjusted for these molecular markers in our multivariable models, due to limited sample size, we were not able to further stratify by these markers. Therefore, our findings warrant additional investigation in future larger-sized studies with sufficient power for these stratified analyses. Sixth, we cannot exclude the possibility of biases related to tumor heterogeneity and contaminated normal cells. In addition, a previous study reported cell-type heterogeneity in LINE-1 methylation levels [42], which might affect our results. However, an experienced pathologist, Dr. Shuji Ogino, carefully reviewed H&E-stained slides of all cases and identified tumor areas in each section, which minimized the possibility of these biases.
Future prospects
Our study has shown a prognostic interaction between smoking and tumor LINE-1 methylation levels measured by the bisulfite-PCR-pyrosequencing method. We used the average of the proportions of C nucleotides at the 4 CpG sites as LINE-1 methylation levels, but DNA methylation may vary in specific repetitive elements of genomes. Recently, several novel approaches have been developed to explore detailed epigenetic profiling. Bock et al. have shown that locus-specific DNA methylation assays in combination with machine learning algorithms can predict global DNA methylation levels more accurately [43]. Subsequently, Zhang et al. reported that a random forest algorithm could accurately predict genome-wide repetitive element methylation using microarray data [44]. Furthermore, nanopore sequencing has enabled us to conduct a direct and real-time analysis of long DNA fragments electronically, which leads to the elimination of amplification bias and efficient assembly, compared to short-read sequencing [45]. These methods can be used in future studies for accurately measuring DNA methylation levels.
Conclusion
Our findings suggest that the association of smoking status at diagnosis with colorectal mortality may be stronger in cases with low LINE-1 methylation level tumors than in those with intermediate or high LINE-1 methylation level tumors. Considering the need for more accurate CRC prognostication, future larger-sized studies are warranted to confirm our findings and guide further exploration into underlying pathways.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are not publicly available due to participant confidentiality and privacy concerns. Further information including the procedures to obtain and access data from the Nurses’ Health Studies and Health Professionals Follow-up Study is described at https://www.nurseshealthstudy.org/researchers (contact email: nhsaccess@channing.harvard.edu) and https://sites.sph.harvard.edu/hpfs/for-collaborators/."
Abbreviations
- CI:
-
Confidence interval
- CIMP:
-
CpG island methylator phenotype
- H&E:
-
Hematoxylin and eosin
- HPFS:
-
Health Professionals Follow-up Study
- HR:
-
Hazard ratio
- IPW:
-
Inverse probability weighting
- LINE-1:
-
Long interspersed nucleotide element-1
- METS:
-
Metabolic equivalent task score-hours
- MPE:
-
Molecular pathological epidemiology
- MSI:
-
Microsatellite instability
- NHS:
-
Nurses’ Health Study
- PCR:
-
Polymerase chain reaction
- SD:
-
Standard deviation
References
Giovannucci E, Colditz GA, Stampfer MJ, et al. A prospective study of cigarette smoking and risk of colorectal adenoma and colorectal cancer in U.S. women. J Natl Cancer Inst. 1994;86(3):192–9.
Giovannucci E, Rimm EB, Stampfer MJ, et al. A prospective study of cigarette smoking and risk of colorectal adenoma and colorectal cancer in U.S. men. J Natl Cancer Inst. 1994;86(3):183–91.
Alwers E, Carr PR, Bandury B, et al. Smoking behavior and prognosis after colorectal cancer diagnosis: a pooled analysis of 11 studies. JNCI Cancer Spectr. 2021;5(5):pkab077.
Wang TH, Hsia SM, Shih YH, et al. Association of smoking, alcohol use, and betel quid chewing with epigenetic aberrations in cancers. Int J Mol Sci. 2017;18(6):1210.
Zeilinger S, Kühnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8(5):e63812.
Wan ES, Qiu W, Baccarelli A, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–82.
Sheaffer KL, Elliott EN, Kaestner KH. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev Res (Philadelphia, Pa). 2016;9(7):534–46.
Hur K, Cejas P, Feliu J, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63(4):635–46.
Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(5):1204–25.e1212.
Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117(9):1847–54.
Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100(23):1734–8.
Akimoto N, Zhao M, Ugai T, et al. Tumor long interspersed nucleotide element-1 (LINE-1) hypomethylation in relation to age of colorectal cancer diagnosis and prognosis. Cancers (Basel). 2021;13(9):2016.
Akimoto N, Ugai T, Zhong R, et al. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol. 2021;18(4):230–43.
Belanger CF, Hennekens CH, Rosner B, Speizer FE. The nurses’ health study. Am J Nurs. 1978;78(6):1039–40.
Rimm EB, Giovannucci EL, Willett WC, et al. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet (London, England). 1991;338(8765):464–8.
Baba Y, Huttenhower C, Nosho K, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125.
Irahara N, Nosho K, Baba Y, et al. Precision of pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa, and peripheral blood cells. J Mol Diagn. 2010;12(2):177–83.
Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105(15):1151–6.
Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9(3):305–14.
Ogino S, Kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8(2):209–17.
Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn. 2005;7(3):413–21.
Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135.
Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–606.
Seaman SR, White IR. Review of inverse probability weighting for dealing with missing data. Stat Methods Med Res. 2013;22(3):278–95.
Liu L, Nevo D, Nishihara R, et al. Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol. 2018;33(4):381–92.
Benjamin DJ, Berger JO, Johannesson M, et al. Redefine statistical significance. Nat Hum Behav. 2018;2(1):6–10.
Zhu Y, Yang SR, Wang PP, et al. Influence of pre-diagnostic cigarette smoking on colorectal cancer survival: overall and by tumour molecular phenotype. Br J Cancer. 2014;110(5):1359–66.
Phipps AI, Shi Q, Newcomb PA, et al. Associations between cigarette smoking status and colon cancer prognosis among participants in North Central Cancer Treatment Group Phase III Trial N0147. J Clin Oncol. 2013;31(16):2016–23.
Jayasekara H, English DR, Haydon A, et al. Associations of alcohol intake, smoking, physical activity and obesity with survival following colorectal cancer diagnosis by stage, anatomic site and tumor molecular subtype. Int J Cancer. 2018;142(2):238–50.
Yang B, Jacobs EJ, Gapstur SM, et al. Active smoking and mortality among colorectal cancer survivors: the Cancer Prevention Study II nutrition cohort. J Clin Oncol. 2015;33(8):885–93.
Swets M, Zaalberg A, Boot A, et al. Tumor LINE-1 methylation level in association with survival of patients with stage II colon cancer. Int J Mol Sci. 2016;18(1):36.
Lou YT, Chen CW, Fan YC, et al. LINE-1 Methylation status correlates significantly to post-therapeutic recurrence in stage III colon cancer patients receiving FOLFOX-4 adjuvant chemotherapy. PLoS One. 2015;10(4):e0123973.
Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science (New York, NY). 2003;300(5618):455.
Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–66.
Hamada T, Nowak JA, Masugi Y, et al. Smoking and risk of colorectal cancer sub-classified by tumor-infiltrating T cells. J Natl Cancer Inst. 2019;111(1):42–51.
Ugai T, Väyrynen JP, Haruki K, et al. Smoking and incidence of colorectal cancer subclassified by tumor-associated macrophage infiltrates. J Natl Cancer Inst. 2022;114(1):68–77.
Ogino S, Nowak JA, Hamada T, et al. Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut. 2018;67(6):1168–80.
Ogino S, Nowak JA, Hamada T, et al. Insights into pathogenic interactions among environment, host, and tumor at the crossroads of molecular pathology and epidemiology. Annu Rev Pathol. 2019;14:83–103.
Mima K, Kosumi K, Baba Y, et al. The microbiome, genetics, and gastrointestinal neoplasms: the evolving field of molecular pathological epidemiology to analyze the tumor-immune-microbiome interaction. Hum Genet. 2021;140(5):725–46.
Phipps AI, Passarelli MN, Chan AT, et al. Common genetic variation and survival after colorectal cancer diagnosis: a genome-wide analysis. Carcinogenesis. 2016;37(1):87–95.
Liu L, Nishihara R, Qian ZR, et al. Association between inflammatory diet pattern and risk of colorectal carcinoma subtypes classified by immune responses to tumor. Gastroenterology. 2017;153(6):1517–30.e14.
Knothe C, Shiratori H, Resch E, et al. Disagreement between two common biomarkers of global DNA methylation. Clin Epigenetics. 2016;8:60.
BLUEPRINT consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34(7):726–37.
Zheng Y, Joyce BT, Liu L, et al. Prediction of genome-wide DNA methylation in repetitive elements. Nucleic Acids Res. 2017;45(15):8697–711.
Ewing AD, Smits N, Sanchez-Luque FJ, et al. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol Cell. 2020;80(5):915–28.
Acknowledgements
We deeply thank hospitals and pathology departments throughout the USA for generously providing us with tissue specimens. In addition, we would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-Up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The authors assume full responsibility for the analyses and interpretation of these data.
Funding
This work was supported by the US National Institutes of Health (NIH) grants (P01 CA87969 to M.J. Stampfer; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; U01 CA167552 to W.C. Willett and L.A. Mucci; R01 CA118553 to C.S.F.; R01 CA169141 to C.S.F.; R01 CA137178 to A.T.C.; K24 DK098311 to A.T.C.; R35 CA197735 to S.O.; R01 CA151993 to S.O.; K07 CA190673 to R.N.; R03 CA197879 to K.W.; R21 CA222940 to K.W. and M.G.; and K07 CA188126 to X.Z.; and R21 CA230873 to S.O. and K.W.; Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17 to C.S.F. and M.G.), administered by the American Association for Cancer Research, a scientific partner of SU2C; Investigator Initiated Grants from the American Institute for Cancer Research (to K.W.); Nodal Award (2016-02) from the Dana-Farber Harvard Cancer Center (to S.O.); and by grants from the Project P Fund, The Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance and SU2C; National Natural Science Foundation of China (81372286) to G.D. Y.C., P.L., and L.L. were supported by a scholarship grant from Chinese Scholarship Council. K.K., K.A., and T.U. were supported by grants from the Overseas Research Fellowship from the Japan Society for the Promotion of Science (JP2017-775 to K.K.; JP201860083 to K.A.; JP201960541 to T.U.). M.G. was supported by a Conquer Cancer Foundation of ASCO Career Development Award. K.F., K.H., and T.U. were supported by fellowship grants from the Uehara Memorial Foundation. T.U. was supported by grants from Mishima Kaiun Memorial Foundation and Prevent Cancer Foundation. K.H. was supported by a fellowship from the Mitsukoshi Health and Welfare Foundation. S.A.V. was supported by grants from the Finnish Cultural Foundation and Orion Research Foundation sr. A.T.C. is a Stuart and Suzanne Steele MGH Research Scholar. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
K.W., R.N., S.O., Y.C., K.I., and T.H. developed the main concept and designed the study. J.K., Y.C., K.H., K.F., L.L., T.U., K.A., N.A., T.H., K.I., K.K., S.S., C.D., P.L., C.G., J.P.V., S.A.V., M.S., X.Z., and D.A.D. were responsible for the collection of tumor tissue and acquisition of epidemiologic, clinical, and tumor tissue data, including histopathological, immunohistochemical, and immunofluorescent characteristics. J.K., Y.C., K.H., K.F., L.L., T.U., R.N., and K.W. performed the data analysis and interpretation. J.K., Y.C., K.H., K.F., L.L., T.U., M.C.L., J.P.V., S.G., G.D., M.G., E.G., A.T.C., C.S.F., S.O., J.A.M., R.N., J.A.N., and K.W. prepared the manuscript and contributed to critical revision for important intellectual contents. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. All participants provided informed consent.
Consent for publication
Not applicable.
Competing interests
A.T.C. previously served as a consultant for Bayer Pharma AG, Pfizer Inc., and Janssen. This study was not funded by Bayer Pharma AG, Pfizer Inc., or Janssen. M.G. receives research funding from Bristol-Myers Squibb and Merck. This study was not funded by Bristol-Myers Squibb or Merck. C.S.F. has been a consultant for Eli Lilly and Co; Entrinsic Health Solutions, Inc; Pfizer Inc; Merck & Co., Inc; Sanofi S.A.; F. Hoffmann-La Roche Ltd; Genentech. Inc; Merrimack Pharmaceuticals, Inc; Dicerna Pharmaceuticals, Inc; Bayer Pharma AG; Agios Pharmaceuticals, Inc; Gilead Sciences, Inc; Five Prime Therapeutics, Inc; and Taiho Pharmaceutical Co, Ltd. C.S.F. is currently employed by Genentech, Roche. J.A.M. has also served as an advisor/consultant to Ignyta, Array Pharmaceutical, and Cota. R.N. is currently employed by Pfizer Inc.; she contributed to this study before she became an employee of Pfizer Inc. This study was not funded by any of these commercial entities. The other authors declare that they have no conflicts of interest.
Use of standardized official symbols
We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products, including BRAF, CACNA1G, CDKN2A, CRABP1, DNMT1, IGF2, KRAS, MLH1, NEUROG1, PIK3CA, RUNX3, SOCS1, and TP53; all of which are described at www.genenames.org. The official symbols are italicized to differentiate from non-italicized colloquial names that are used along with the official symbols. This format enables readers to familiarize themselves with the official symbols for genes and gene products together with common colloquial names.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
Frequencies between smoking status and pack-years among past/current smokers.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kishikawa, J., Ugai, T., Fujiyoshi, K. et al. Smoking and colorectal cancer survival in relation to tumor LINE-1 methylation levels: a prospective cohort study. Epigenetics Commun. 2, 4 (2022). https://doi.org/10.1186/s43682-022-00012-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43682-022-00012-y