Abstract
Background
Detecting the genetic and physiological variations in two Japanese quail strains could be used to suggest a new avian model for future breeding studies. Consequently, two estimations were performed on two Japanese quail strains: gray quail strain (GJQS) and white jumbo quail strain (WJQS). The first estimation was conducted on carcass characteristics, breast muscles, breast concentration of collagen type I, and body measurements. In contrast, blood samples were collected for the second estimation for genomic DNA extraction and genetic analysis.
Results
A total of 62 alleles out of 97 specific alleles (63.92%) were detected overall loci (14 microsatellite loci) for the two strains. A total of 27 specific alleles of WJQS were observed, and 35 were obtained for GJQS. The percentage of similarity was 48.09% ranged from 4.35 with UBC001 to 100% with GUJ0051. WJQS had greater body weights and a higher value of pectoral muscle and supracoracoideus muscle than GJQS. The breast muscles of GJQS exhibited a higher concentration of type I collagen than the WJQS. Furthermore, males showed higher concentrations of collagen type I than females. WJQS showed a higher body length, chest girth, chest length, thigh length, thigh girth, drumstick length, and drumstick girth (cm) than GJQS. WJQS showed more significant differences in carcass traits compared with GJQS.
Conclusion
The physiological differences between WJQS and GJQS were ascertained with microsatellite markers, which indicated high polymorphism between these strains. These observations provided a scientific basis for evaluating and utilizing the genetic resources of WJQS and GJQS in a future genetic improvement program.
Similar content being viewed by others
Background
Japanese quail (Coturnix coturnix japonica) is currently the smallest poultry species reared primarily for meat and egg production [1]. It has unique characteristics, including rapid growth, quick life cycle, disease resistance, early sexual maturity, high rate of lay, and lower feed consumption [2, 3]. These characteristics significantly differ between the Japanese quail strains. Therefore, quails were divided into different strains according to breeding, either meat production quails, egg production quails, or dual-purpose quails. Besides, Mohammed et al. [4] reported various plumage color mutations in Japanese quails and white and gray plumage colors that may be considered different quail strains. Thus, it is essential to assess the existence of genetic and substantial physiological variations within these strains to establish effective breeding programs to improve the most important economic traits. Many studies have reported some estimates of genetic parameters for various traits of Japanese quail’s body and performance [5, 6]. They concluded that the continuous increase could improve the growth performance and egg production of the Japanese quail in their genetic potential and favorable management conditions. Hence, the characterization of indigenous bird populations’ physiological parameters and genetic diversity is a prerequisite tool for providing needed information for the conservation of useful genotypes to improve efficiency and significant productivity of birds [7, 8]. The mentioned characterization can be achieved using the microsatellite marker technique to estimate the variability and genetic relationships between and within the bird’s populations [9]. Habimana et al. [10] also evaluated the degree of genetic diversity and phylogenetic relationships between IC populations in Rwanda by using simple sequence repeats markers. Therefore, the purpose of this study was to characterize and detect the genetic and physiological variations in two strains of Japanese quails (gray quail strain (GJQS) and white jumbo quail strain (WJQS)) and to determine the molecular description for these strains by physiological measurements and molecular genetics, and lastly to suggest a new avian model for future breeding studies.
Methods: birds’ husbandry and ethics
Two strains of Japanese quails (GJQS n = 60 and WJQS n = 62), at 5 weeks old, were maintained at the quail experimental farm of the Biological Application Department, Nuclear Research Center, Egypt. Birds were reared in battery cages of 100 × 60 × 50 cm (length × width × height) in size, categorized by each strain, and fed a diet matching with the National Research Council [11]. All procedures used in this investigation were approved by the scientific and ethics committee of the Biol. Appli. dep., (protocol number 187; date of approval: 28 August 2019), according to the policies and guidelines of the institutional poultry care and use committee.
Collection of data
Two estimations were performed on GJQS and WJQS as follows:
Physiological estimations
Bird weights and biometric body measurements (cm) were collected individually for each strain using a flexible measuring tape. The biometric body measurements (cm) include body length, chest length, chest girth, thigh length, thigh girth, drumstick length, and drumstick girth. Birds were slaughtered, and the empty carcass, liver, heart, intestine, gizzard, proventriculus, and spleen were weighed, recorded, and expressed as a percentage of live body weight. Dressing percentage and carcass yield were estimated as described by Abd El-Moneim et al. [12] and Abdel-Moneim et al. [13]. The breast muscle was exposed, and both right and left supracoracoideus (SC) and pectoralis major muscle (PC) were excised, weighed, and expressed in absolute weight and relative weight. Samples of the breast muscle tissue 0.1 g were taken out and rinsed with 1x PBS, freeze-thaw cycles to break the cell membranes, and centrifuged for 5 min at 5000×g, 2–8 °C. The supernatant was removed, and the quantitative determination of collagen type I concentrations were determined immediately using the ELISA kit (catalog number csb-e0804r) produced by CUSABIO TECHNOLOGY LLC (http://www.cusabio.com), Houston, TX 77054, USA.
Genetic analysis
Blood samples were collected from GJQS (n = 20) and WJQS (n = 22), for genomic DNA extraction according to methods described by Sambrook et al. [14] as follows: a half milliliter of the blood sample was withdrawn from the jugular vein on EDTA tube as anti-coagulant (0.2 ml of 0.5 M EDTA). DNA was freshly extracted from the whole collected EDTA-blood. Two and a half milliliter of lysis buffer TSTM (20 mM Tris-HCl pH 7.6, 640 mM sucrose, 2% Triton X-100, 10 mM MgCl2) was added to the aliquot. The mixture was centrifuged, and the pellet was suspended in 150 μl proteinase K, 1.5 ml nuclei lysis buffer, and 110 μl SDS 20%. After overnight incubation at 37 °C, the proteins were removed by NaCl 6 M, and the DNA was precipitated by ice-cold absolute ethanol.
Microsatellite genotyping: source of primers
Fourteen primer pairs of microsatellite markers, as shown in Table 1 were designed according to the literature of Kayang et al. [15], Charati et al. [16], Moradian et al. [17], and Roushdy and El-Sayed [9]. Applying these locations specifically in the present study will explain the results of physiological estimations such as body weight, morphometric body measurements, carcass traits, breast muscle weight, and soluble collagen.
Polymerase chain reaction
The PCR was performed using 50–100 ng genomic DNA in a 25 μl reaction volume containing 10 μl Master Mix (Emerald AMP GT PCR Master Mix, Takara Bio. Inc. composed of 10 pmol of each primer, DNA polymerase, optimized reaction buffer, dNTPs and a density reagent). The premix also contained a vivid green dye, which is separated into blue and yellow dye fronts. The PCR reactions were carried out under the following conditions: an initial denaturation step (for 4 min at 95 °C), followed by 35 cycles of denaturation (for 1 min at 95 °C), annealing (at 48–64 °C for 1 min) at optimized primer annealing temperature (Table 1), and then extension (for 1 min at 72 °C) and final extension (for 10 min at 72 °C). Amplified fragments were analyzed on 10% polyacrylamide gel and stained with Ethidium bromide. The gels were photographed, and images were analyzed using the Gel Documentation System (Alpha imager TM 2200, Cell Biosciences).
Statistical analysis
The physiological results were analyzed with the general linear model and variance procedure analysis between quail strains using the statistical software [18]. Tukey’s procedure for multiple comparison tests was used to identify significant differences of values at a significance level of 5%. All scored microsatellite data were firstly corrected to estimate each allele size according to its number of repeats for each marker. All possible extracted species’ figures were carried out employing an Arlequin 3.5 software package after data conversion using the CONVERT program. The POPGENE software package [19] was used to calculate allele frequencies, observed (HO) and expected (HE) heterozygosities, and ENA for WJQS and GJQS.
Results
Physiological estimation
Body measurements
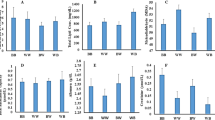
Differences in body measurements between two Japanese quail strains are presented in Table 2. The WJQS showed a higher value in body length, chest girth, chest length, thigh length, thigh girth, drumstick length, and drumstick girth (cm) than GJQS. No significant differences were observed inside the sex strain in the mentioned body measurements.
Body weight and carcass characteristics
Variations between two Japanese quail strains in marketing body weight and carcass characteristics are recorded in Table 3. The WJQS had a larger body (312.7.0 vs. 279.3 g, P ˂ 0.001) weights compared with GJQS. The relative weight of carcass yield, dressing, liver, heart, proventriculus, and spleen except intestine and gizzard were significantly higher in WJQS than GJQS. Moreover, sex differences were observed inside strain itself, whereas male quail showed significant values in dressing, heart, and carcass yield percentages, while female quail showed significant values in marketing body weight, liver, intestine, proventriculus, and spleen percentages.
Breast muscle characteristics
The investigation of the breast muscle characteristics and collagen content of breast muscle in two Japanese quail strains (white vs. gray) is illustrated in Table 4. The results of WJQS indicated higher weight values for the right pectoralis major muscle (PC) (23.39 vs. 20.37 g, P 0.004), left PC (23.31 vs. 20.07 g, P 0.004), right supracoracoideus muscle (SC) (7.29 vs. 5.83 g, P ˂ 0.001), and left SC (8.1 vs. 5.57 g, P ˂ 0.001) than the GJQS. Furthermore, sex differences were observed inside strain; females of WJQS showed a higher value of SC and PC muscles than males, while males of GJQS showed a higher value of SC and PC muscles than females.
Genetic estimations
Microsatellite loci, annealing temperatures, primer sequence, gene bank accession numbers, repeat array, and band size are shown in Table 1. Annealing temperatures ranged from 48 with UBC001 to 64 with GUJ0053; the band size ranged from 35 to 610 bp in WJQS and GJQS with fourteen microsatellite markers, as shown in Table 1.
The total number of alleles was 97 out of fourteen microsatellite markers ranged from 3 with GUJ0013 and GUJ0051 to 23 with UBC001 in WJQS and GJQS. The total number of alleles per strain was 62 ranged from one in GUJ0048 to 12 in UBC001 with a mean of 4.43 in WJQS while, the total number of alleles was 70 ranged from two in GUJ0028, GUJ0053, and GUJ0054 to 12 in UBC001 with a mean of 5.00 in Japanese quails strain as shown in Table 5. Regarding specific alleles, a total of 62 out of 97 alleles (63.92%) were detected overall loci (14 microsatellite loci) versus two strains. For WJQS, 27 with a mean of 1.93 specific alleles were observed, while 35 with a mean of 2.50 were obtained for GJQS. These specific alleles would be utilized as a strain fingerprint in WJQS and GJQS.
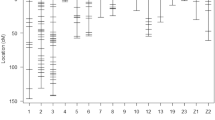
Specific alleles for the WJQS strain could not be detected using the markers GUJ0013, GUJ0021, GUJ0048, GUJ0051, GUJ0087, UBC002, and UBC005. Also, GUJ0051, GUJ0053, and GUJ0099 produced no specific alleles for Japanese strain, as shown in Table 5. In respect to ENA, it was used to corollary detect the expected heterozygosity (HE) where the effective number of alleles is the highest when heterozygosity is high. In our results, the lowest ENA was 1.35 for GUJ0053 when HE was 0.26 while the highest ENA was 17.98 for UBC001 when HE was 0.96 (Table 5 and Fig. 1). The degree of genetic variation of the microsatellite loci was reflected by heterozygosity in strains. Also, high heterozygosity indicated a high genetic diversity as well as a high degree of genetic variation. Out of 14 microsatellite sequences selected for detecting the differentiation and similarity between WJQS and GJQS, the percentage of similarity was 48.09% ranged from 4.35 with UBC001 to 100% with GUJ0051. The highest number of alleles per strain, the specific alleles, the total number of alleles, and the significant number of alleles were detected in UBC001, which had 12 numbers of alleles per strain and 11 specific alleles within the two strains and a total number of 23 alleles with ENA was 17.98, and the lower percentage of similarity was 4.35.
The estimated proportions of WJQS for each individual are represented by the green bar’s length, as shown in Fig. 2. The red bars in the group indicate that several hybrids and probably even pure Japanese individuals (whole red bar) are present in the GJQS.
Allelic frequencies were calculated based on all fourteen microsatellite loci. The highest allele frequency overall loci were 1.00 for allele 80 at GUJ0048 locus in WJQS, while the lowest one (0.023) was for allele 65 at GUJ0051 locus in WJQS. Also, the highest average of allele frequency estimated was 0.33 at loci GUJ0013 and GUJ0053. Meanwhile, the lowest one was 0.04 at locus UBC001.
Polymorphic information content (PIC) refers to the possibility that a progeny acquires some allelic markers from its father or mother, describing the variation degree of microsatellite loci. The value of PIC for WJQS ranged from 0.19 to 0.89 in GUJ0053 and UBC001, with a mean of 0.58 in WJQS. While it ranged from 0.32 to 0.89 in GUJ0053 and UBC001 with a mean of 0.62 in GJQS as shown in Table 6, these differences reflect high genetic variability between two quail strains.
In this study, three markers GUJ0013 (0.47), GUJ0051 (0.49), and GUJ0053 (0.19) were reasonably informative (0.50 > PIC>0.25). Marker of GUJ0048 (0.00) was a slightly informative marker, and most of the loci were highly informative with WJQS. Four markers GUJ0021 (0.43), GUJ0028 (0.42), GUJ0051 (0.47), and GUJ0053 (0.32) were reasonably informative (0.50 > PIC>0.25), while the majority of the loci were highly informative (PIC ≥ 0.50) with GJQS. The analysis of molecular variance estimated by the Arlequin 3.5 software package as standard genetic strain input data is presented in Table 7. Variance components proved that most genetic diversity obtained in the current study is represented within individuals (24.11%) rather than others. Fixation indices give an idea about the strain’s structure in terms of straining coefficient and strain differentiation. Strain fixation indices traced a 0.759 variation, referring to differences among individuals versus total variance (FIT). While among strains, differences versus total variance were the lowest fixation indices (FST = 0.178), indicating a low level of strain differentiation. These observations might be explained as approximate equality of the average total number of alleles detected for each strain overall loci. It was 4.43 for WJQS and 5.00 for GJQS, as shown in Table 7.
Discussion
Physiological estimation
The significant differences in the two-color variants studied (WJQS and GJQS) in the marketing body weight and different body measurements such as body length, chest girth, chest length, thigh length, thigh girth, drumstick length, and drumstick girth reflect the differences between WJQS and GJQS in body sizes and shape, indicating positive relationships between body weights and body measurements (Tables 5 and 6). Moreover, the obtained results confirm the physiological variations between WJQS and GJQS, which may be due to the existence of genetic variation between them. The obtained results agree with several workers that reported a positive correlation between live body weight and morphometric body measurements in Isa Brown and Ilorin ecotype chickens [20], in two commercial broiler strains [21], in Japanese Quails [22], in the French broiler guinea fowl [23], and two commercial meat-type chickens [24]. Moreover, it is well-known that body weight is considered the most important physiological indices for evaluating different livestock species for numerous reasons, including its relation with body growth and other physiological traits such as body morphometric measurements, carcass characteristics, and breast meat yield. In an overall comparison of two quail strains, the WJQS attained greater physiological parameters in terms of body weight, carcass yield, most of the body organs, and breast meat yield than GJQS. This might be attributed to the superior genetic potential of WJQS than GJQS, which lead to higher marketing bodyweight and produced more massive carcass and more meat. These observations are consistent with Ojedapo et al. [21] and Ahmad et al. [22] who reported a strong genetic correlation between body weight and carcass traits. Similarly, other studies [25,26,27] reported higher carcass yield in selected heavy lines of Japanese quail superior to that of a non-selected.
Furthermore, it is generally accepted that both PC and SC muscles are positively correlated with marketing body weight, muscle mass, and meat quality. In this mention, Młynek et al. [28] showed that dressing percentage significantly affected carcass and pectoralis significant muscle weight and soluble collagen. Therefore, the selection for increased live body weight in earlier studies performed by Ryu et al. [29] and Rehfeldt et al. [30] was a suitable way to enhance Japanese quail’s growth performance. Baylan et al. [31] reported a similar finding; Anjum et al. [32] observed a higher breast meat yield in birds selected for body weight. In another explanation, Choi et al. [33] reported a positive correlation between DNA contents and muscle weights between quail lines. The present study’s results sustain this finding regarding the collagen content of breast muscle in WJQS and GJQS. Significant differences were found: breast muscles of GJQS exhibited a higher concentration of type I collagen, almost three-fold than the WJQS (529.2 vs. 187.8 Pg/ml, P < 0.001), respectively (Table 7). Furthermore, sex differences were observed inside each strain; the male showed a higher value of collagen type I concentration than the female (437.2 vs. 279.8 Pg/ml, P < 0.001), respectively. It is well-known that intramuscular collagen is an essential parameter to the meat industry; an increased amount of this component may affect toughness and meat quality. In other words, the most abundant fibrous form of collagen in muscle is type I, which considers the main structural protein of connective tissues present in meat, providing meat toughness and rigidity and involved in the structural integrity and several physiological functions [34, 35]. Moreover, the significant factor affecting meat tenderness is the maturity of connective tissues, which is a function of chemical cross bonding of the collagen in the muscle, which increases with age; hence, the tough meat is found in older birds [36]. Therefore, the differences in collagen content in WJQS and GJQS, which feed on the same diet and under the same age, may confirm the existence of genetic variations between them.
Genetic estimations
Examining the results of the 14 microsatellite markers in this work showed some genetic differences in two quail strains WJQS and GJQS. As a result, the observed genetic differences confirmed the presence of physiological variations between WJQS and GJQS, such as body weight, carcass characteristics, body measurements, breast muscle weights, and collagen type I concentration of breast muscle. Our results are consistent with similar studies conducted by Charati et al. [16] and Moradian et al. [17], which showed the relation of these locations with cold carcass weight, breast meat weight, and body dimensions and carcass parameters. The obtained results on annealing temperatures and the size of bands in two strains (WJQS and GJQS) with 14 microsatellite markers agree with Roushdy and El-Sayed’s [9] results from 60 to 470 bp with UBC001, UBC002, UBC005, and GUJ0028. Also, it agrees with Kayang et al. [15], wherein the values ranged from 96 to 284 bp. Besides, the mentioned obtained values could be informative for such studies, according to Kawahara-Miki et al. [37], who suggested that the allele sizes of the DNA fragments for the 101 markers ranged from 7 to 36 repeats and 91 to 311 bp, respectively, in the Japanese quail, while Bai et al. [38] observed that the annealing temperatures ranged from 46 to 58 with 12 microsatellite markers. Moreover, the total number of alleles per strain agreed with Kayang et al. [15], who reported that the average of 1.9 alleles per locus ranged from one to four alleles. Also, Choi et al. [39] reported that the mean number of alleles in each breed ranged from 3.59 to 6.63. Further studies carried on different quail genotypes by Bai et al. [40], Kawahara-Miki et al. [37], Bai et al. [38], Shimma and Tadano [41], and Habimana et al. [10] reported the allele size of 48, 70, 197, 308, and 305, respectively. Furthermore, the specific alleles for WJQS were 27 and 35 with GJQS. These values were lower than those observed by Roushdy and El-Sayed [9], who detected 68 out of 136 specific alleles (50%) overall loci (12 microsatellite loci) versus two species. Also, Habimana et al. [10] showed 20% of private alleles. A high value of heterozygosity (51.91%) between two quail strains with 14 microsatellite loci and the effective number of alleles that ranged between 1.6504 (MCW0078) and 8.901 (LEI0234) indicated the relatively rich genetic variation of two strains and a significant genotype of WJQS than GJQS. However, ENA’s obtained value was more significant than the estimated value reported by Habimana et al. [10]. On the other side, the allele frequency over all loci ranged from 0.023 to 1.00 with WJQS. These results agree with El-sayed [42], who reported that the specific allele frequency value ranged from 0.05 to 0.50 based on 15 microsatellite loci used for Fayoumi and Dandarawi breeds. When PIC values were examined, it was seen that a substantial portion of working locus markers provided information at a high level. When Table 3 was analyzed in terms of PIC means, the value was highly informative (PIC ≥ 0. 50); it was observed that there was a difference among quail strains. According to the classification of Botstein et al. [43], the highly informative markers have PIC values > 0.50, the reasonably informative markers have a PIC value between 0.25 and 0.50, and the slightly informative markers have PIC value less than 0.25. In this study, three markers GUJ0013 (0.47), GUJ0051 (0.49), and GUJ0053 (0.19) were reasonably informative (0.50 > PIC > 0.25). Marker of GUJ0048 (0.00) was a slightly informative marker. The majority of the loci were highly informative with WJQS. Four markers GUJ0021 (0.43), GUJ0028 (0.42), GUJ0051 (0.47), and GUJ0053 (0.32) were reasonably informative (0.50 > PIC > 0.25), while the majority of the loci were highly informative (PIC ≥ 0. 50) with GJQS. This suggests that a high degree of polymorphism has potentially been maintained in two strains WJQS and GJQS. Also, Bai et al. [38] reported that the average PIC of 12 microsatellite markers at Chinese yellow quail, Chinese black quail, and Korean quail which are 0.6853, 0.6401, and 0.6565 respectively were highly informative (PIC ≥ 0.50). Habimana et al. [10] showed that the PIC ranged from reasonably to highly informative since the PIC for the loci MCW0103 and LEI0234 were 0.3488 and 0.8775, respectively. Fixation indices give an idea about the strain’s structure in terms of straining coefficient and strain differentiation. Also, the investigation had been done by Vargas et al. [44] who reported that FIS ranged from a minimum of − 0.034 (MCW014) to a maximum of 0.727 (MCW014) with an average of 0.146 (0.1254–0.1638). Finally, Habimana et al. [10] showed that the contribution of 28 microsatellites for population segregation (determined by FST statistics) varied from 0.000 (MCW0037) to 0.158 (ADL0268).
Conclusion
This study showed highly physiological differences between WJQS and GJQS in live body weight, carcass characteristics, body measurements, breast muscle weights, and collagen type I concentration of breast muscle. These physiological variations were ascertained with selected 14 microsatellite markers, which indicated the relatively rich genetic variation of the two strains and a significant genotype of WJQS than GJQS. These results succeeded in introducing a scientific basis for the evaluation and utilization of genetic resources of WJQS and GJQS in the next breeding programs for genetic improvement of the breed in an attempt to stop the continuous inbreeding system in quail farming and, consequently, improve the production performance of Japanese quail.
Availability of data and materials
Not applicable
Abbreviations
- PIC:
-
Polymorphism information content
- PCR:
-
Polymerase chain reaction
- HO:
-
Observed heterozygosities
- HE:
-
Expected heterozygosities
- ENA:
-
Effective number of alleles
- FIS:
-
Fixation indices (among strains)
- FST:
-
Fixation indices (among individuals within strains)
- FIT:
-
Fixation indices (within individuals)
- IC:
-
Inbreeding coefficient
- D.F:
-
Degrees of freedom
- S.S:
-
Sum of squares
- GGA, CJA, and QL:
-
Linkage group chicken and Japanese quail chromosome
References
Faraji-Arough H, Rokouei M, Maghsoudi A, Ghazaghi M (2018) Comparative study of growth patterns in seven strains of Japanese quail using nonlinear regression modeling. Turk J Vet AnimSci 42:441–451
Jatoi AS, Sahota AW, Akram M, Javed K, Jaspal MH, Hussain J, Mirani AH, Mehmood S (2013) Effect of different body weight categories on the productive performance of four close-bred flocks of Japanese quail (Coturnix coturnix japonica). J Anim Plant Sci 23(1):7–13
Nasar A, Aminoor Rahman MD, Nazmul Hoque MD, Talukder AK, Das ZC (2016) A survey of Japanese quail (Coturnix coturnix japonica) farming in selected areas of Bangladesh. Veterinary World 9(9):940–947
Mohammed AF, Nasr MR, Ali MAH (2017) Performance, carcass traits, meat quality and amino acid profile of different Japanese quails strains. J Food Sci Technol 54(13):4189–4196
Minvielle F, Gourichon D, Ito S, Inoue-Murayama M, Riviere S (2007) Effects of the dominant lethal yellow mutation on reproduction, growth, feed consumption, body temperature, and body composition of the Japanese quail. Poult. Sci 86:1646–1650
Sruoga A, Slavenaite S, Butkauskas D (2007) Application of microsatellite primers for investigation of Peking ducks hybrid line, Proc. of the 13th Baltic animal straining conference, Parnu, pp 20–23
Davila SG, Gil MG, Resino-Talavan P, Campo JL (2009) Evaluation of diversity between different Spanish chicken strains, a tester line, and a White Leghorn population based on microsatellite markers. Poult Sci 88:2518–2525
Kayang S, Lumineau A, Vignal JM, Boutin Rognon X (2010) Evidence for introgressive hybridization of wild common quail (Coturnixcoturnix) by domesticated Japanese quail (Coturnix japonica) in France. Conserv.Genet 11(3):1051–1062
Roushdy KH, El-Sayed MA (2017) Genetic assessment of two chicken strains and two Japanese quail lines by using chicken and quail microsatellite markers reciprocally. Egypt Poult Sci 37(I):21–36
Habimana R, Okeno TO, Ngeno K, Mboumba S, Assami P, Gbotto AA et al (2020) Genetic diversity and population structure of indigenous chicken in Rwanda using microsatellite markers. PLoS One 15(4):e0225084
National Research Council (1994) Nutrient requirements of poultry, 9th revised edition. The National Academies Press, Washington, DC
Abd El-Moneim AE, El-Wardany I, Abu-Taleb AM, Wakwak MM, Ebeid TA, Saleh AA (2020) Assessment of in ovo administration of Bifidobacterium bifidum and Bifidobacterium longum on performance, ileal histomorphometry, blood hematological, and biochemical parameters of broilers. Probiotics Antimicrob Proteins 12(2):439–450
Abdel-Moneim AE, Sabic EM, Abu-Taleb AM, Ibrahim NS (2020) Growth performance, hemato-biochemical indices, thyroid activity, antioxidant status, and immune response of growing Japanese quail fed diet with full-fat canola seeds. Tropl Anim Health Prod 52(4):1853–1862
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn 3 volumes. Cold Springs Harbor Laboratory, Cold Springs Harbor
Kayang B, Miho Inoue-Murayama B, Hoshi T, Matsuo K, Takahashi H, Minezawa M, Makotomizutani SI (2002) Microsatellite loci in Japanese quail and cross-species amplification in chicken and Guinea fowl. Genet Sel Evol 34:233–253
Charati H, Esmailizadeh AK, Jabbari R, Ori H, Moradiyan SS (2015) Genetic mapping of quantitative trait loci affecting skeletal architecture in Japanese quail. Vet Res 8(4):45–52
Moradian H, Nasirifar E, Sohrabi SS, Esmailizadeh AK (2016) Microsatellite mapping of quantitative trait loci affecting carcass traits on chromosome 1 in half-sib families of Japanese quail (Coturnix japonica). J Livestock Sci Technol 4(1):53–60
SPSS Inc (2010) PC + statistics. ver. 18.0. IBM Corp, Armonk
Yeh FC, Boyle T, Rongcai Y, Ye Z, Xian JM (1999) POP gene, version 1.31. A Microsoft window based freeware for population genetic analysis. University of Alberta, Edmonton
Fayeye TR, Hagan JK, Obadare AR (2014) Morphometric traits and correlation between body weight and body size traits in Isa Brown and Ilorin ecotype chickens. Iranian J Appl Anim Sci 4(3):609–614
Ojedapo LO, Amao SR, Aderibigbe DO (2016) Evaluation of the parameters needed to describe the growth traits of two commercial broiler strains. J Nat Sci Res 6(4):96–101
Ahmad S, Mehmood S, Javed K, Mahmud A, Usman M, Rehman A, Ishaq HM, Hussain J, Ghayas A (2018) Different selection strategies for the improvement of the growth performance and carcass traits of Japanese quails. Braz J of Poultry Sci 20(3):497–506
Dzungwe JT, Gwaza DS, Egahi JO (2018) Statistical modeling of body weight and body linear measurements of the French broiler Guinea fowl in the humid tropics of Nigeria. Poult Fish Wildl Sci 6:2
Amao SR (2019) Estimation of body weight from linear body measurements in two commercial meat-type chickens raised in southern Guinea environment of Nigeria using principal component analysis approach. Int J Nutr Agric Res 6(2):39–46
Akram M, Hussain J, Sahota AW, Ahmad S, Jatoi AS, Ali A, Dawood M (2012) Genetic gain in 4 week body weight through mass selection in 4 close-bred stocks of Japanese quail. Book of abstracts.32nd Congress of Zoology, Lahore.
Hussain J, Akram M, Sahota AW, JavedK AHA, Mehmood S, Ahmad S, Sulaman R, RabbaniI JAS (2013) Selection for higher three week body weight in Japanese quail: 1. Effect on growth performance. J Anim Plant Sci 23(6):1496–1500
Khaldari MA, Pakdel H, MehrabaniYegane A, NejatiJavaremi BP (2010) Response to selection and genetic parameters of body and carcass weights in Japanese quail selected for 4- week body weight. Poult Sci 89:1834–1841
Młynek K, Charuta A, Janiuk I, Ole A, Głowińska B (2016) Effect of dressing percentage on chemical composition, microstructure and quality traits of pectoralis major muscle in female Japanese quail. Europ Poult Sci 80
Ryu YC, Rhee MS, Kim BC (2004) Estimation of correlation coefficients between histological parameters and carcass traits of pig longissimus dorsi muscle. Asian Austral J Anim Sci 17:428–433
Rehfeldt C, Henning M, Fiedler I (2008) Consequences of pig domestication for skeletal muscle growth and cellularity. Livest Sci 116:30–41
Baylan M, Canogullari S, Sahin A, Copur G, Baylan M (2009) Effect of different selection methods for body weight on some genetic parameters. J Anim Vet Adv 8:1385–1391
Anjum M, Sahota AW, Akram M, Javed K, Mehmood S (2012) Effect of selection on productive performance of desi chicken for four generations. J Anim Plant Sci 22(1):1–5
Choi YM, Shin S, Wick MP, Choe JH, Lee K (2013) Muscle fiber characteristics of pectoralis major muscle as related to muscle mass in different Japanese quail lines. Animal 7(10):1665–1670
Heino J (2007) The collagen family members as cell adhesion proteins. Bioessays 29(10):1001–1010
Maiorano G, Sobolewska A, Cianciullo D, Walasik K, Elminowska-Wenda G, Slawínska A, Tavaniell S, Zylinska J, Bardowski J, Bednarczyk M (2012) Influence of in ovo prebiotic and synbiotic administration on meat quality of broiler chickens. Poult Sci 91:2963–2969
Mir NA, Rafiq A, Kumar F, Singh V, Shukla V (2017) Determinants of broiler chicken meat quality and factors affecting them: a review. J Food SciTechnol 54(10):2997–3009
Kawahara-Miki R, Sano S, MitsuoNunome TS, TakehitoKuwayama ST, Kawashima T, Matsuda Y, Yoshimura T, Kono T (2013) Next-generation sequencing reveals genomic features in the Japanese quail. Genomics 101:345–353
Bai JY, Huang Y, Zhang XH, Yang YB, Pang YZ, Qi YX (2018) Polymorphism analysis of 3 quail groups by using microsatellite marker. Life Sci J 15(2) http://www.lifesciencesite.com
Choi SS, Seo JH, Han J-M, Seong J, Lee JH, Kong HS (2019) The development of multiplex PCR microsatellite marker sets for Korean chicken breeds. Int J Poult Sci 18:492–498
Bai JY, Pang Z, Wu Y, Yu SJ, Zhang MQ, Zhao XH, Xu HW (2013) Polymorphism analysis of Chinese yellow quail using microsatellite markers. J Anim Plant Sci 23(4):1072–1076
Shimma K, Tadano R (2019) Genetic differentiation among commercial lines of laying-type Japanese quail. J Poult Sci 56(1):12–19
El-Sayed MA (2012) Determining some genetic loci of productive traits in some indigenous chicken strains using microsatellite marker. Ph.D thesis, Department of Poultry Production, Faculty of Agriculture, Ain Shams University.
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Vargas PAT, León JM, Ortega LRF, Martinez A, Gavilanes AAV, Delgado JV, Landi V (2019) Deciphering the patterns of genetic admixture and diversity in the Ecuadorian creole chicken. Animals 9:670
Acknowledgements
The authors would like to express sincere thanks and deep appreciation to the National Gene Bank, Animal Genetic Resources Dept., Agric. Res. Center, Giza, Egypt, and the Biological Application Department, Nuclear Research Center, Egyptian Atomic Energy Authority, for their cooperation.
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
NSI and AEA participated in Japanese quail sample collection and performed two strains of Japanese quails (gray and white jumbo quails), maintained at the quail experimental farm and the data discussion, data analyses. MAE and HAMA participated in microsatellite analyses of strains (gray and white jumbo quails) and the data discussion and data analyses, and AE participated in drafting of the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The authors declare that all procedures used in this investigation were approved by the scientific and ethics committee of the Boil. Appli. Dept., (protocol number 187; date of approval: 28 August 2019) according to the policies and guidelines of the institutional poultry care and use committee.
Consent for publication
Not applicable
Competing interests
Not applicable
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ibrahim, N.S., El-Sayed, M.A., Assi, H.A.M. et al. Genetic and physiological variation in two strains of Japanese quail. J Genet Eng Biotechnol 19, 15 (2021). https://doi.org/10.1186/s43141-020-00100-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-020-00100-3