Abstract
Background
This study was conducted to detect the presence of T. gondii in milk and blood samples using three different assays: enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), and loop-mediated isothermal amplification assay (LAMP). Whole blood, serum, and milk samples were collected from goats (n = 156), sheep (n = 261), and camels (n = 108) in different governorates in Egypt from December 2019 to February 2021 and screened by ELISA for anti-Toxoplasma IgG antibodies before DNA extraction. The target T. gondii DNA gene was detected and evaluated using the LAMP assay compared to PCR.
Results
T. gondii antibodies were found in milk and serum samples at the rates of (29.26%) and (36.58%) in camels, (34.18%) and (35.89%) in sheep, and (33.7%) and (36.36%) in goats, respectively. Similar to PCR, the percentages of LAMP tests for the detection of the T. gondii DNA gene in milk and blood samples of camels, sheep, and goats were (4.8, 14.63), (6.83, 7.69), and (7.79, 9.09), respectively. LAMP's sensitivity for detecting T. gondii in milk and blood samples, which was identical to that of PCR, was 100%.
Conclusions
The findings clearly demonstrated that there were no variations in T. gondii detection capabilities in milk and blood samples from various animals using both PCR and LAMP tests. It provides a quick, precise, and sensitive method of detecting T. gondii in a variety of samples that may be used both in the field and in laboratory diagnosis.
Similar content being viewed by others
1 Background
T. gondii infection is prevalent worldwide in both animals and humans [1]. The definitive hosts of this parasite are domestic and wild felids while its intermediate hosts are mammals and birds [2]. Cats spread oocysts in the environment where they contaminate food and water with oocysts and tachyzoites that infect intermediate hosts such as livestock and humans [3, 4]. Previous studies have reported the possibility of the presence of tachyzoites in the milk of goats and sheep [5,6,7]. Toxoplasma gondii is a major cause of abortion in farm livestock [8]. Toxoplasmosis can be diagnosed using a variety of methods, including ELISA, latex agglutination tests, and molecular techniques [9,10,11]. However, serological methods have limitations in detecting specific antibodies in early infection, and IgM is not detected during the reactivation of re-infection [12]. Although polymerase chain reaction (PCR) is a sensitive method, it is not used in clinical laboratory diagnosis due to the high cost of materials and the time required for judging results by gel electrophoresis. Therefore, there is still a need to develop a simple, rapid, and cost-effective method of detecting T. gondii in different samples of humans and animals [13]. Serological tests are preliminary tests that detect parasites, while conventional polymerase chain reaction (PCR) is the gold standard for the detection of T. gondii in biological samples from animals because its high sensitivity and reliability. However, the major disadvantage of these methods is that they are quite time-consuming and do not provide quantitative results. The real-time PCR assay is the technique of choice for T. gondii diagnosis and is preferred to conventional PCR. The rapidity, simplicity, and ease of application of results of immunochromatographic tests (ICTs), coupled with the fact that no special equipment is required, make ICT suitable for the field application of T. gondii in different biological samples [14, 15]. The real-time PCR (qPCR) technique is well known for its concurrent detection and quantification of extracted T. gondii–DNA gene amplicon. This technique has significantly shortened time and has comparable performance to cPCR, which is machine-dependent, often expensive, and requires regular maintenance.
The LAMP assay is one of the molecular methods used in various fields for the diagnosis and identification of infectious agents. It is a promising assay that has highly specific and sensitive amplification of DNA to detect infectious agents [16]. Our study aimed to optimize the LAMP assay for the detection and identification of T. gondii DNA in blood and milk samples of camels and small ruminants.
2 Methods
2.1 Samples and study area
A total of 525 blood samples were collected in two vacationer tubes (one with EDTA anticoagulant for molecular assays and the other without anticoagulant for serum collection) from lactating (n = 235), non-lactating (n = 201), and male (n = 89) camels, sheep, and goats from December 2019 to February 2021 in different governorates (Beni Suef, Giza, Monufia, Alexandria, Sharqia, Matruh, and Faiyum) in Egypt. Blood samples were collected from the jugular vein and left at room temperature for 30 min to separate serum samples from the clotted blood cells then centrifuged at 3000 rpm for 10 min to separate serum. After centrifugation, each serum sample was transferred into plastic freezing vials with leak-proof screw caps. Sera were separated, labeled, and kept at − 20 °C until they were examined serologically for T. gondii infection. However, whole blood samples with EDTA were stored at -20 °C until the DNA used for molecular diagnosis was extracted from them. Similarly, milk samples (5 ml to 10 ml) were collected from lactating female camels, sheep, and goats (n = 235) under sterile conditions; teats were cleaned and wiped prior to sample collection in a sterile single-use plastic bottle and immediately transported at 4 °C to the laboratory in a cool box with ice packs. The sample was divided into two parts and kept at a temperature of 20 °C until it was used. The frozen milk samples were thawed at room temperature and centrifuged at 3000 rpm for 10 min. The clear fraction was examined for anti-T. gondii antibodies.
2.2 Positive and negative controls
The Toxoplasma gondii RH strain was kindly obtained from a colony maintained in the Zoonotic Disease Department, Veterinary Research Division (VRD), and National Research Centre (NRC). Also, positive and negative control serum samples were kindly provided by the Parasitology and Animals Diseases Department, NRC, Egypt.
2.3 ELISA assay
The previously collected serum and milk samples were prepared and screened for anti-T. gondii antibodies as described in the manual kit (Enzyme Immunoassay Test Kit, Bio, USA), and modified by [17]. Optimal dilutions for ELISA were adjusted using checkerboard titrations for sera, antigens, and conjugates. Polystyrene plates were coated with 100 μl of antigen/well (soluble antigens extracted from the laboratory-prepared-mice-propagated T. gondii RH strain) at 10 mg/ml in coating buffer and then incubated overnight at 4 °C. The plates were washed three times with phosphate buffered saline (PBS) containing 0.05% Tween 80 (PBS Tween) and blocked for 45 min at 37 °C with carbonate buffer (pH 9.6) containing 10% BSA in a humid chamber. Following three washes, 100 ul of serum or milk sample diluted 1:100 in PBS was added to the plate, which was then incubated for 1 h at 37 °C. After washing as above, 100 ul of HRP (horseradish peroxidase)-protein A conjugate (Mark-Germany) diluted 1:3000 in blocking buffer was added to the wells and incubated at 37 °C for 1 h. After washing, 100 ul of the substrate (0.4 mg/mL of O-phenylenediamine (OPD), 0.0125% hydrogen peroxide in 0.1 M sodium citrate buffer, pH 5.0) was added and left for 10 to 15 min at room temperature in a dark place. The reaction was stopped with 100 ul per well of 0.5 M sulfuric acid and then read for optical density (OD) at 490 nm in a microplate ELISA reader. All samples were run in triplicates. Each plate contained three positive and three negative reference serum samples. The samples were considered positive when their OD was greater than or equal to the cutoff value. The positive samples were subjected to confirmation by PCR and LAMP.
2.4 DNA extraction
The extraction of the genomic DNA of T. gondii from milk and uncoagulated blood samples of different animals was carried out using the technique proposed by [18]. The DNA/RNA extraction kit (iNtRON, Korea) was used according to the manufacturer’s instructions. DNA concentration and purity were measured using the technique by [13]. Prior to DNA extraction, concentration was carried out by centrifugation at 6000 rpm for 10 min. To avoid interference by casein, 1 ml of pellets was treated with 200 μl TE [1 mM EDTA, 10 mM Tris–HCl (pH = 7.6)] and 300 μl 0.5 M EDTA (pH = 8), after which it was suspended and centrifuged at 6000 rpm for 10 min. Somatic cells were diluted in 200 μl of PBS, and DNA was extracted from both blood and milk somatic cells using the DNA extraction kit (iNtRON) according to the manufacturer’s instructions. The DNA purity was measured according to the method described by [13]. The extracted DNA was stored at − 20 °C until it was used. Toxoplasma DNA from tachyzoites and tissue cysts was extracted from RH Toxoplasma tachyzoites, which were counted using a hematocytometer, and, afterward, used in a serial limited dilution method from 1 × 105 to 1 × 10−1 tachyzoite. Also, the RH T. gondii strain was used as a positive control, and sterile deionized water was used as a negative control. The DNA pellet was dissolved in 50 μl of TE and stored at − 20 °C until it was used.
2.5 PCR amplification
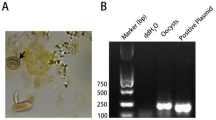
The Toxoplasma gondii B1 gene was amplified using primer sets designed from genes (5OB1/F 5′-GGAACTGCATCCGGTTCATGAG-3′ and 2 OB1/R 5′-TCTTTAAAGCGTTCGTTC-3′) [5]. The following reaction mixture (25 μL) was added to a PCR tube and DNA amplification was done in a final volume of 20 μl (10 μl of 2 × PCR master mix, 1 μl of each primer, 2 μl template DNA (25 ng), and up to 6 μl of nuclease-free water were mixed in a PCR tube). Amplification was performed as follows: denaturation (95 °C/2 min) and 40 cycles consisting of denaturation (95 °C/1 min), annealing (55 °C/30 s), and extension (72 °C/45 s) and final extension (72 °C/10 min). The PCR product was evaluated by electrophoresis at 80 V/15 min and finally examined with a UV transluminator, and a 100 bp DNA ladder was used as a marker.
2.6 LAMP reaction for detection of the DNA
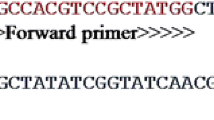
The LAMP reaction was performed according to the method proposed by [19]. We used four primers for the LAMP assay, targeting six conserved regions of T. gondii DNA as shown in Table 1. The (F3): outer forward primer, (B3): outer backward primer, (FIP): forward inner primer and backward inner primer (BIP) are designed by the Primer Explorer Program (http://primerexplorer.jp/elamp4.0.0/index.html). Two sets of each of the four primers are listed in Table 1. We performed LAMP in a total of 25-μL containing primer mix (1μL) that contained 40 pmol of FIP and BIP each, 20 pmol of F3 and B3 each, 12.5 μL of the reaction buffer (New England Biolabs), 2μL of DNA, and 6.5 μL of nuclease-free water. The mixture was incubated at 66 °C for 1 h before being heated to 80 degrees Celsius for 5 min.
2.7 Analytical sensitivity and specificity of the LAMP assay
The primer sequences were selected to specifically amplify the AF and/or RE primer sequence for the T. gondii genome as described previously. The AF or RE was diluted tenfold serially as 109 copies to 1 copy to determine the sensitivity of the LAMP assay. Besides, tachyzoites of T. gondii RH strain harvested from mice were counted on a hemocytometer and diluted as 30, 25, 20, 15, 10, 5, and 1 tachyzoites RH strain in 400 μL of PBS. For the LAMP assay, genomic DNA was extracted from the 400-μL sample solution with a DNA extraction kit. We used T. gondii -DNA positive strains (strains RH), as well as positive T. gondii–DNA from Tissue cyst Toxoplasma RH, Toxoplasma RH (tachyzoites) mice, Toxoplasma (tachyzoites) bovine, Toxoplasma First, in a reaction containing Tissue s cyst Toxoplasma RH, Toxoplasma RH (tachyzoites) mice, Toxoplasma (tachyzoites) bovine, Toxoplasma (tachyzoites) small ruminants, Toxoplasma (tachyzoites) equines, and Toxoplasma RH (tachyzoites) SPF-DNA, National Research Centre, Veterinary Research Institute, Egypt. DNA extracted from E. coli, Staphylococcus spp, Salmonella species, Giardia lamblia, N. caninum (provided from the Parasitology and Animals Diseases Department, NRC, Egypt) has also been used to prove the specificity of the Toxoplasma LAMP assay.
2.8 Statistical analysis
The Statistical Package for the Social Sciences (SPSS) (version 17.0; SPSS, Inc, USA) was used to analyze the collected data. The McNemar test was used to compare the sensitivity of the LAMP assay with PCR. Receiver operating characteristic (ROC) curve analysis and area under the curve (AUC) were used to determine clinical sensitivity and specificity.
3 Results
The rates of Toxoplasma gondii detection in sheep, goats, and camels' lactating female serum samples in the different governorates (Beni Suef, Giza, Monufia, Alexandria, Sharqia, Matruh, and Faiyum) in Egypt by ELISA were 42/117 (35.89%), 28/77 (36.36%), and 15/41 (36.58%), while the rates of T. gondii detection in milk samples were 2/41 (4.8%), 8/117 (6.83%), and 6/77 (7.79%), respectively. The prevalence of T. gondii was higher in male goats than in sheep; 18/31 (58%) and 15/31 (48.3%), respectively. In lactating and non-lactating females, the rates were 18/48 (37.5%) and 45/113 (33.98%), respectively, as illustrated in Figs. 2 and 3. The rates of T. gondii antibodies byELISA were 5/27 (18.51%), 14/40 (35%), and 15/41 (36.58%) of male, non-LF, and LF of camel serum samples as shown in Fig. 1.
3.1 Specificity of the LAMP reaction
The designed four LAMP primers in Table 2 were used to amplify the AF and/or RE fragment(s) from extracted DNA from parasites, including E. coli, Staphylococcus spp, Salmonella species, Giardia lamblia, N. caninum, Tissue Oocyte Toxoplasma RH, Toxoplasma RH (tachyzoites) mice, and Toxoplasma (tachyzoites) bovine. First, in a reaction containing Tissue cyte Toxoplasma RH, Toxoplasma RH (tachyzoites) mice, Toxoplasma (tachyzoites) bovine, Toxoplasma (tachyzoites) small ruminants, Toxoplasma (tachyzoites) equines, and Toxoplasma RH (tachyzoites) SPF-DNA, clear changes in their viable colors were seen. While using DNA from other protozoa (E. coli, Staphylococcus spp, Salmonella species, Giardia lamblia, N. caninum) and negative control SPF with PBS pH 7.2, negative control proteineal fluid from mice inoculated with PBS as the template, no such target was observed. It is suggested that the LAMP assay with target-specific primers showed high specificity in the detection of T. gondii. Table 2 shows that the designed four LAMP primers were used to amplify the AF and or RE fragment from extracted DNA from parasites, including E. coli, Staphylococcus spp, Salmonella species, Giardia lamblia, N. caninum, Tissue Oocyte Toxoplasma RH, Toxoplasma RH (tachyzoite) mice, Toxoplasma (tachyzoites) bovine and negative control SPF with PBS pH 7.2, negative control proteineal fluid from mice inoculated with PBS as the template, no such target was observed. It is suggested that the LAMP assay with target-specific primers showed high specificity in the detection of T. gondii.
3.2 LAMP assay sensitivity
The LAMP assay's sensitivity was tested by detecting DNA amplification with various copies ranging from 101 to 106. The sensitivity of LAMP was also tested using genomic DNA isolated from various quantities of tachyzoites as the template. When genomic DNA from at least 5 tachyzoites was used, the amplified DNA fragments were visible. Although there was no DNA product in the condition with only one tachyzoite's genome DNA, the amplified target segment did not show any visible change in reactions. According to the results, the LAMP assay was very sensitive in detecting Toxoplasma even with only 10 copies of the target fragment or five tachyzoites in the sample. A total of 525 clinical samples from apparently normal animals were subjected to examination by cPCR and LAMP assays. After analyzing the samples using the PCR assay, 63 (12%) and 462 (88%) of the samples were found to be positive and negative, respectively, for T. gondii, as shown in Table 3. Also, the cPCR using the B1 gene primer sequence products of the 63 positive samples showed 100% identity. Then, both the previous positive and negative samples were analyzed by the LAMP assay, with different primers, the AF and/or RE primer, AF-LAMP, and RE-LAMP. The previous 63 positive samples were all proven to be positive for the Toxoplasma AF-LAMP and RE-LAMP assays as shown in Tables 2 and 3. So, the detection rate using LAMP primers was identical to that obtained using conventional PCR. While positive PCR results (15%, 15%, and 20%) were obtained in male, non-LF, and LF blood samples collected from camels, similar results were obtained using the LAMP assay, which examined the same samples as shown in Table 2 and Fig. 4.
Detection of T. gondii in milk and blood samples by LAMP. Positive reaction turned green under UV light after the addition of a fluorescent green dye in tubes, from left to right, respectively: negative control. LAMP assay reaction under daylight without UV light after the addition of the fluorescent dye
Table 3 clearly shows that the prevalence of T. gondii in sheep and goat milk samples was (26, 40%). While T. gondii antibodies were detected in 12/41 (29.26%) of the examined camel milk samples as shown in Figs. 1, 2, and 3, the seropositive samples were subjected to further examination using PCR and LAMP, and the DNA of T. gondii was found to be 2/41 (4.8%) in camel milk, 8/117 (6.83%) in sheep milk, and 6/77 (7.79%) in goat milk as in shown in Table 4.
3.3 A comparison between LAMP and conventional PCR was conducted
Conventional PCR and LAMP tests were used to examine 525 clinical samples from different animals that were apparently normal. After cPCR analysis, 63 (12%) and 462 (88%) of the samples were determined to be positive and negative, respectively, for T. gondii, as shown in Tables 2 and 3. The LAMP assay was used to examine positive and negative samples using different primers (AF-LAMP and RE-LAMP). Toxoplasma AF-LAMP and RE-LAMP tests were identical to cPCR results as in (Table 4). The results of examining 235 blood and milk samples from different animals by cPCR were 22 (9.36%) and 16 (6.8%), indicating that the detection rate by LAMP primers was identical to that obtained using conventional PCR as shown in Table 4.
4 Discussion
Toxoplasmosis is an important zoonotic parasitic disease transmitted from fields to any animal or human through the contamination of pasture with T. gondii oocytes of infected cats’ faces [7]. T. gondii oocytes shed from cat's faces from 4 to 14 days after infection [20]. Goats and sheep feed on green pastures contaminated with T. gondii oocytes. The present study revealed that the high incidence of T. gondii in lactating goats’ and lactating sheep's milk, but not in lactating camel milk, is due to the difference between the types of pastures of small ruminates and the camels in different governorates as shown in Figs. 1, 2, and 3. These findings are in line with those of [16]. It is reported that the camels feed mainly on the dry matter that is present in the desert, and this feed fairway is contaminated by cats’ feces, which is the main source of Toxoplasma oocytes. In the present study, the prevalence of T. gondii in serum samples of male and female camels, sheep, and goats was determined by ELISA. The rates of contamination, which were 15/31 (48.3%), and 18/31 (58%), were higher in males than in lactating and non-lactating females, 18/48 (37.5%), and 45/113 (39.8%), respectively. The rates of T. gondii antibody detection in the serum samples of camels were 5/27 (18.51%), 14/40 (35%), and 15/41 (36.58%) in males, non-LF, and LF, respectively, in different governorates in Egypt as shown in Figs. 1, 2, and 3. The prevalence reported in our study is slightly lower than the one found by Saad et al. [17]. The prevalence rates in sheep and goats are lower than those detected in two separate studies (64.34% and 64.9%, respectively) conducted in 2009 [18,19,20]… Seropositivity in goats was found to be higher than in sheep, and the selective feeding habits of goats may be explained the high seroprevalence detected in this species. The prevalence identified in a similar study in goats in another country (75%) is significantly higher than the one we found in goats in our study [21].
The rates of T. gondii DNA detection by PCR in blood samples of camels were 15%, 15%, and 20% in males, non-LF, and LF, and similar results were obtained using the LAMP assay. These results are shown in Table 2 and are similar to those obtained by [21]. The results found similar or slightly higher rates than those [22], who reported the seroprevalence of T. gondii in camels’ serum and milk samples. They reported that the seroprevalence of T. gondii in camels’ serum and milk samples was similar to that reported by [23], who stated that the prevalence of T. gondii antibodies in camel's milk was 3.12%. Also, they explained that the highest incidence of T. gondii occurrence in milk in goats and sheep, but not in camels, is due to the difference in the types of pastures of small ruminants that feed on green pastures contaminated by T. gondii oocytes. Camels feed in the desert dry fodder fairway of cats, which is the main source of Toxoplasma oocytes [24].
The prevalence of T. gondii in sheep and goat milk samples by ELISA was 40/117 (34.18%) and 26/77 (33.7%). While T. gondii antibody was detected in 12/41 (29.26%) of the examined camel milk samples, as shown in Table 3 and Figs. 1, 2, and 3, the positive samples were subjected to further examination using PCR for detection of T. gondii DNA in milk samples of different animals and found 2/41 (4.8%) in camel milk, 8/117 (6.83%) in sheep milk, and 6/77 (7.79%) in goat milk, which can be explained by the lower incidence of T. gondii DNA detected by PCR in comparison to T. gondii antibodies may be due to the fact that IgG antibodies are produced late in the infection, and the parasite is localized in the organs and tissues rather than circulated in the blood; so, it does get to the milk. These findings, which are presented in Tables 3 and 4, are consistent with those of previous studies [25, 26]. The molecular assays are more accurate, sensitive, and specific than serological techniques. LAMP was sensitive, simple, and fast for DNA amplification; it took less than 1 h under isothermal conditions at 66 °C. The LAMP method has been proven to be very specific for the detection of T. gondii. Moreover, the LAMP method was much more sensitive in detecting T. gondii than other methods, and it was also much cheaper. We evaluated the specificity of the LAMP assay by screening T. gondii genomic DNA from other parasites. No cross-reactivity with E. coli, Staphylococcus spp, Salmonella species, Giardia lamblia, or N. caninum DNA samples was detected. The detection limit of this LAMP assay was five tachyzoites, which was similar in sensitivity to the PCR target at the B1 gene as shown in Tables 2 and 3. Also, it used several primers that recognize six to eight regions of the target DNA that eliminate the nonspecific binding of primers and are easily detected from the negative amplifications without the need for any gel electrophoresis to judge results [27].
In the present study, we detected T. gondii DNA in milk and blood samples of different animals by LAMP. The positive reaction medium turned green under UV light after a fluorescent dye was added as shown in Fig. 4. The results were obtained without the need for any gel electrophoresis or gel-documentation system, which reduces the amount of time required to evaluate the results. While the LAMP reaction tube under daylight without UV light gave white turbidity in the end product, this may be due to the magnesium pyrophosphate produced during the reaction, which leads to the production of a visible white precipitate. These results and explanations are in line with those of previous studies [11, 16]. The obtained results revealed that the incidence of Toxoplasma gondii antibodies as detected by ELISA in sheep milk samples was 39.66%. Lower values were reported by many authors who showed that the prevalence of Toxoplasma gondii was 5.94% in ovine milk samples by an ELISA test in Iran, and they found that the prevalence of anti-Toxoplasma gondii IgG antibodies in the raw milk of sheep was 3.76%.
The results shown in Tables 3 and 4 demonstrate that the LAMP assay is highly sensitive in the detection of T. gondii DNA in blood samples (100%), is relatively easy, and does not require advanced technical skills or expensive equipment. This result is in agreement with those of previous studies [11, 19]. However, LAMP seems to be extremely sensitive to contamination; as such, it should be performed in a biological safety cabinet type I as a separate part for LAMP. The LAMP assay (AF-LAMP and RE-LAMP) and cPCR using the B1 gene primer are similar, and the LAMP assay was carried out according to the reported general reaction conditions. Several samples were judged as positive via our LAMP assay, indicating surprisingly high sensitivity and specificity values, which are by far the most similar to those of conventional PCR at 100%, as shown in Tables 3 and 4. In this study, T. gondii DNA was detected in camel milk samples at 2/41 (4.8%), the lowest detection rate of DNA in milk samples relative to whole blood samples among different animal species, indicating that all seropositive animals do not secrete T. gondii tachyzoites into their milk, which could be a limiting factor for PCR results as in Table 4, and this result is in agreement with the results of previous studies [22, 28]. The high seropositivity and the presence of parasite DNA in milk led to the conclusion that T. gondii infection is present in the sheep population and that there is a risk of the human population becoming infected due to the consumption of raw, natural milk. Hill and Dubey [17] found Toxoplasma gondii DNA in 6.05% and 2.7% of the raw goat milk samples using the PCR technique in Brazil. The prevalence of Toxoplasma gondii DNA in naturally infected milk samples n in Italy was 13% [17]. However, Egyptian researchers estimated that Toxoplasma gondii DNA was present in 8% of goats from rural areas in Sharkia, Egypt. Meanwhile, a higher prevalence was recorded; the prevalence of Toxoplasma gondii DNA was 32.56% in goat milk from a farm in Slovakia. These variations in results may be attributed to the immune statuses of the animals, the timing of the infection, and the genetic compositions of the host and the organism. Also, the explanation is in line with those of several authors [17] who reported that the rate of occurrence of tachyzoites in the milk of naturally infected sheep was 6.5% of T. gondii DNA in milk samples. Observing turbidity with the naked eye or adding intercalating dyes to the reaction tube after amplification is generally used to identify LAMP findings. However, these procedures need some expertise or the use of UV light to evaluate the outcome. Products may be easily spotted by visual examination when using LAMP products, reducing the reaction time and allowing the assay to be utilized for field-testing. Furthermore, because it is introduced before amplification, the LAMP test reduces the likelihood of false positives due to aerosol contamination [22, 28]. As a result, the LAMP method proposed here looks to be a potential alternative to cPCR for T. gondii detection; hence, this approach appears to be a potential alternative to cPCR for the identification of T. gondii DNA in blood samples (100 percent) in various species. T. gondii DNA was identified in blood and milk samples, as reported by previous studies [11,12,13]. However, for the examination of blood and milk samples, LAMP and cPCR findings were in perfect agreement. Because blood appears to be the transport channel for tachyzoites between body tissues, molecular tests are valuable for the early diagnosis of T. gondii in asymptomatic animals. Finally, the LAMP assay can be considered as one of the most accurate, specific, sensitive, and rapid diagnostic assays for early T. gondii tachyzoite detection in milk and blood samples.
5 Conclusions
Toxoplasmosis is a major public health problem and continues to be a risk to human and animal health due to its presence in animal milk and milk byproducts. The prevalence of T. gondii by ELISA was higher in males than in females of camels and small ruminants that represent a source of Toxoplasma infection, so animals should be examined by serological tests to help in monitoring and controlling of infection spreading. LAMP assay only requires a water bath or heat-block. Also, visual detection of amplification products, superior or similar to PCR, can be an alternative assay for other diagnostic methods. Finally, the LAMP assay is a promising, simple, cheap, and rapid diagnostic tool for the diagnosis of active T. gondii in different animals' samples.
Availability of data and materials
Not applicable.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- ELISA:
-
Enzyme-linked immunosorbent assay
- LAMP:
-
Loop-mediated isothermal amplification assay
- LF:
-
Lactating female
- M:
-
Male
- Non-LF:
-
Non-lactating female (dry or young female)
- PCR:
-
Polymerase chain reaction
- T. gondii :
-
Toxoplasma gondii
References
Abdalhamed AM, Hassanain MA, Zeedan GSG, Shaapan RM (2019) Evaluation of Toxoplasma gondii propagated in specific pathogen free embryonated chicken egg, for diagnosis of toxoplasmosis in equids and human. J Parasit Dis 43(3):498-505. https://doi.org/10.1007/s12639-019-01117-3
Aguirre AA, Longcore T, Barbieri M, Dabritz H, Hill D, Klein PN, Lepczyk C, Lilly EL, McLeod R, Milcarsky J (2019) The one health approach to toxoplasmosis: epidemiology, control, and prevention strategies. EcoHealth 16(2):378–390. https://doi.org/10.1007/s10393-019-01405-7
Al-Malki ES (2021) Toxoplasmosis: stages of the protozoan life cycle and risk assessment in humans and animals for an enhanced awareness and an improved socio-economic status. Saudi J Biol Sci 28(1):962–969. https://doi.org/10.1016/j.sjbs.2020.11.007
Azimpour-Ardakan T, Fotouhi-Ardakani R, Hoghooghi-Rad N, Rokni N, Motallebi A (2021) Designing and developing of high-resolution melting technique for separating different types of Toxoplasma gondii by analysis of B1 and ROP8 gene regions. J Microbiol Methods 184:106188. https://doi.org/10.1016/j.mimet.2021.106188
Dehkordi FS, Haghighi Borujeni MR, Rahimi E, Abdizadeh R (2013) Detection of Toxoplasma gondii in raw caprine, ovine, buffalo, bovine, and camel milk using cell cultivation, cat bioassay, capture ELISA, and PCR methods in Iran. Foodborne Pathog Dis 10(2):120–125
Fallahi S, Tabaei SJS, Pournia Y, Zebardast N, Kazemi B (2014) Comparison of loop-mediated isothermal amplification (LAMP) and nested-PCR assay targeting the RE and B1 gene for detection of Toxoplasma gondii in blood samples of children with leukaemia. Diagn Microbiol Infect Dis 79(3):347–354
Ferens WA, Hovde CJ, Underwood WJ, Blauwiekel R, Delano ML, Gillesby R, Song Y, Xu YT, Altamirano FLG, Barr JJ (2019) News on therapeutic management of MDR-tuberculosis: a literature review. Front Microbiol 6:96. https://doi.org/10.14202/vetworld.2018.355-362
Fernández-Escobar M, Calero-Bernal R, Benavides J, Regidor-Cerrillo J, Guerrero-Molina MC, Gutiérrez-Expósito D, Collantes-Fernández E, Ortega-Mora LM (2020) Isolation and genetic characterization of Toxoplasma gondii in Spanish sheep flocks. Parasites Vectors 13(1):1-14
Ghoneim NH, Shalaby SI, Hassanain N, Zeedan GSG, Soliman Y, Abdalhamed AM (2010) Comparative study between serological and molecular methods for diagnosis of toxoplasmosis in women and small ruminants in Egypt. Foodborne Pathog Dis 7(1):17–22. https://doi.org/10.1089/fpd.2008.0223
Ghoneim NH, Shalaby SI, Hassanain NA, Zeedan GSG, Soliman YA, Abdalhamed AM (2009) Detection of genomic toxoplasma gondii dna and anti-toxoplasma antibodies. Life Sci J 6:54–60
Guowu Z, Kai Z, Xifeng W, Chunhui J, Chengcheng N, Yue Z, Jun Q, Qingling M, Xingxing Z, Kuojun C (2020) Occurrence of gastrointestinal parasites in camels in the Tianshan Mountains pastoral area in China. J Vet Res 64(4): 509. https://doi.org/10.2478/jvetres-2020-0071
Hegazy MK, Awad SI, Saleh NE, Hegazy MM (2020) Loop mediated isothermal amplification (LAMP) of Toxoplasma DNA from dried blood spots. Exp Parasitol 211:107869. https://doi.org/10.1016/j.exppara.2020.107869
Hill DE, Dubey JP (2013) Toxoplasma gondii prevalence in farm animals in the United States. Int J Parasitol 43(2):107–113. https://doi.org/10.1016/j.ijpara.2012.09.012
Johnson JD, Butcher PD, Savva D, Holliman RE (1993) Application of the polymerase chain reaction to the diagnosis of human toxoplasmosis. J Infect 26(2):147–158. https://doi.org/10.1016/0163-4453(93)92788-X
Kamani J, Mani AU, Egwu GO (2010) Seroprevalence of Toxoplasma gondii infection in domestic sheep and goats in Borno state. Nigeria Trop Anim Health Prod 42(4):793–797. https://doi.org/10.1007/s11250-009-9488-3
Khan AH, Noordin R (2020) Serological and molecular rapid diagnostic tests for Toxoplasma infection in humans and animals. Eur J Clin Microbiol Infect Dis 39(1):19-30. https://doi.org/10.1007/s10096-019-03680-2
Kumari S, Kumar RR, Mendiratta SK, Kumar, Dhananjay, Kumar A, Jawla J, Rana P, Kumar (2021). Development of loopmediated isothermal method and comparison with conventional PCR assay for rapid on spot identification of tissue of cattle origin. J Food Sci Technol 58:4608–4615. https://doi.org/10.1007/s13197-020-04948-8
Luptáková L, Petrovová E (2013) Diagnostics of Toxoplasma gondii in milk samples of sheep from eastern Slovakia. Anim Welfare Ethol Hous Syst 9:567–569
Mancianti F, Nardoni S, D’Ascenzi C, Pedonese F, Mugnaini L, Franco F, Papini R (2013) Seroprevalence, detection of DNA in blood and milk, and genotyping of Toxoplasma gondii in a goat population in Italy. BioMed Res Int 2013. https://doi.org/10.1155/2013/905326
Mosa IS, Saheb EJ, Mahdi SG, Abdul-Karim MI, Khistawi AN (2017) An epidemiology study of some protozoan parasitic diseases in Iraq from 2011 Till 2015. Al-Nahrain J Sci 20(3):115–120
Pleyer U, Gross U, Schlüter D, Wilking H, Seeber F (2019) Toxoplasmosis in Germany: epidemiology, diagnosis, risk factors, and treatment. Dtsch Arztebl Int 116(25):435–444. https://doi.org/10.3238/arztebl.2019.0435
Rostami A, Karanis P, Fallahi S (2018) Advances in serological, imaging techniques and molecular diagnosis of Toxoplasma gondii infection. Infection. 46(3):303–315. https://doi.org/10.1007/s15010-017-1111-3
Saad NM, Hussein AAA, Ewida RM (2018) Occurrence of Toxoplasma gondii in raw goat, sheep, and camel milk in Upper Egypt. Veterinary World 11(9):1262–1265
Sadek OA, Abdel-Hameed ZM, Kuraa HM (2015) Molecular detection of Toxoplasma gondii DNA in raw goat and sheep milk with discussion of its public health importance in Assiut Governorate. Assiut Vet Med J 61(145):166–177
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd ed., Vols 1, 2 and 3
Shaapan RM (2016) The common zoonotic protozoal diseases causing abortion. J Parasit Dis 40(4):1116–1129. https://doi.org/10.1007/s12639-015-0661-5
Sharma S, Sandhu KS, Bal MS, Kumar H, Verma S, Dubey JP (2008) Serological survey of antibodies to Toxoplasma gondii in sheep, cattle, and buffaloes in Punjab. India J Parasitol 94(5):1174–1175
Vismarra A, Barilli E, Miceli M, Mangia C, Bacci C, Brindani F, Kramer L (2017) Toxoplasma gondii and pre-treatment protocols for polymerase chain reaction analysis of milk samples: a field trial in sheep from Southern Italy. Ital. J food Saf 6(1):6501. https://doi.org/10.4081/ijfs.2017.6501
Acknowledgements
We would like to acknowledge National Research Centre, Dokki, Egypt, for facilities and funds Project No # 12010138 # during this work.
Funding
This work funded by Project No# 12010138 # National Research Centre fund (NRC).
Author information
Authors and Affiliations
Contributions
Dr. AMA, Dr.GZ, and Dr. RMS designed the plan of work for the experiment and carried out the laboratory work. Dr. AE participated in the laboratory work and in drafting the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical statement the study was approved ethically by the Medical Research Ethical Committee, National Research Centre, Egypt, under registration number # 19- 148 #.
Consent for publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeedan, G.S.G., Abdalhamed, A.M., Shaapan, R.M. et al. Rapid diagnosis of Toxoplasma gondii using loop-mediated isothermal amplification assay in camels and small ruminants. Beni-Suef Univ J Basic Appl Sci 11, 1 (2022). https://doi.org/10.1186/s43088-021-00184-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43088-021-00184-x