Abstract
Background
Infection control in cystic fibrosis (CF) patients plays a crucial role in improving the survival of patients with CF. Antimicrobial sensitivity patterns in these patient groups in our country are currently lacking. Therefore, the purpose of the study was to evaluate the microbiological cultures and antimicrobial susceptibility pattern of pediatric CF patients.
Method
A total of 50 respiratory samples were prospectively collected from the period between February 2021 and October 2021. Sputum and oropharyngeal swabs were processed for culture and microbiological testing. Sample collection and evaluation were performed according to the Good Laboratory Practice guidelines (GLP). Informed written consent was ensured before participation. Statistical analysis was performed with SPSS v 26.
Result
The median age of the children was 30 months (6–120) months, with a male predominance (66% vs 34%). Single and two organisms were isolated in 72% (n = 36) and 12% (n = 6) of cases, respectively. During the study period, 36% of the patients harbored Pseudomonas aeruginosa, 18% harbored Klebsiella pneumoniae, and both Staphylococcus aureus and Escherichia coli were detected in 16% of cases. Levofloxacin was found to be the most active antibiotic agent with 100% susceptibility. In contrast, nearly all isolates were resistant to amoxicillin, erythromycin and rifampicin.
Conclusion
Levofloxacin is the most effective agent to treat CF patients. Active surveillance of the resistance pattern should always continue to be promoted.
Similar content being viewed by others
Background
Repeated bacterial lung infection leads to the progressive development of respiratory incompetency in patients with cystic fibrosis (CF), resulting in expanded mortality and morbidity [1, 2]. In cystic fibrosis, mutation of the CFTR gene impairs innate defenses, such as the mucociliary clearance system and bactericidal activity of the airways, resulting in accelerated pulmonary colonization and chronic infection with harmful bacteria [3]. Near one-third of patients with CF experience lung problems since their early childhood and develop radiological evidence of bronchiectasis [4]. Hemophilus influenzae and Staphylococcus aureus are the dominant causative agents in the early stage of the disease, but with the progression of age and disease, Pseudomonas aeruginosa becomes the most prevalent and notorious for CF lungs [5, 6]. In addition, other opportunistic microbes, including Stenotrophomonas maltophilia, Achromobacter spp., and Burkholderia cepacia, are also responsible for exacerbations of pulmonary symptoms [7, 8]. Moreover, as the patient ages, frequent use of antibiotics facilitates antimicrobial resistance along with alteration of microbiome diversity in the affected lung milieu. Thus, classical microbes are being replaced by newly emerged pathogens, and benign colonizers are becoming the culprits of disease advancement [7, 9, 10]. Therefore, considering such dynamic microbiology, it is often difficult to control CF airway infection [11]. However, during an exacerbation, to regain optimum pulmonary efficacy, there is no alternative of suitable antibiotics, and it should be under the guidance of an in vitro antibiotic susceptibility test (AST) [12].
On the other hand, it is currently not a disease of the West; rather, it is fairly reported from different corners of the world, with an incidence ranging from 1:2000 to 1:35,000 in new births. Despite limited comprehensive documentation of CF cases in Bangladesh, approximately 100 cases have been reported in the last decade [11, 13,14,15,16]. To establish standard care of these infection-prone children, knowing the microbiological profile and antibacterial resistance pattern is therefore essential. Therefore, studying the microbiological pattern and susceptibility to antibiotics among the collected samples of children with CF was the objective of the study.
Materials and methods
This observational study was carried out over a period between February 2021 and October 2021 among the patients admitted to the inpatient department of Dhaka Shishu Hospital, a tertiary level hospital in Bangladesh designated to care for the children. Approval from the institutional review board (IRB) was sought before the commencement of the study. Written informed consent was obtained from the parents of the affected child. Patients were included in this investigation on conditions in which they had CF, authenticated by their measured sweat chloride level ≥ 90 mmol/L (performed on SANASOL (SM-01) sweat analyzer, Sanasol Ltd. Hungary) along with characteristic clinical features [17]. Children with suggestive clinical features of asthma, aspiration pneumonia, pulmonary tuberculosis, foreign body aspiration, primary immunodeficiency, congenital heart disease, pulmonary abscess, congenital diaphragmatic hernia, and congenital lobar emphysema were excluded from this study.
Parents were interviewed, and a structured questionnaire was employed to fill out for all cases. Complete blood counts (CBCs), random blood sugar (RBS), sputum and oropharyngeal swab cultures with antibiograms, Mantoux tests (MTs), and chest X-rays (CXRs) were performed in all patients. Other supporting investigations, such as stool for fat droplets, serum albumin level, liver function test (LFT), and echocardiography, were also performed. Reviewing of the previous medical records was also performed.
Specimen collection and culture
Sputum samples were collected into sterile containers provided by the laboratory, screwed tightly, and labeled. Parents were directed to keep their child refrain from food for 1 h before collection. Subjects who were unable to spontaneously expectorate oropharyngeal swabs were collected with the assistance of parents or by asking the child to open the mouth, gently rubbing a sterile swab stick over the pharyngeal wall and replacing that in a sterile plastic container.
Within 1 h of collection, samples were transported to the microbiology laboratory of the same institution. Samples were inoculated on Mac-Conkey’s agar, chocolate agar, and blood agar and incubated at 37 °C for 24 h. After completion of 24 h, colonies were identified, subcultured, and subjected to biochemical reactions for identification of microorganisms as per standard technique [18].
Antimicrobial susceptibility test
Susceptibility to antimicrobial agents of all isolates was determined by the Kirby Bauer modified disc diffusion technique using Mueller–Hinton agar plates, and zones of inhibition were interpreted according to Clinical and Laboratory Standards Institute (CLSI), 2017 guidelines. Antibiotic discs such as amoxicillin, ceftazidime, cefepime, vancomycin, piperacillin, imipenem, meropenem, linezolid, cotrimoxazole, gentamicin, amikacin, erythromycin, ciprofloxacin, levofloxacin, rifampicin clindamycin, tobramycin, timentin, and aztreonam were used, and proper concentrations of the drugs were maintained [19].
Ethics statement
The institutional review board (IRB) of Bangaldesh Institute of Child Health, Dhaka, Bangladesh approved the study protocol. Memo no: [BICH-ERC-5–2-21].
Statistical analyses
Statistical analyses were conducted using SPSS software: v-26. Continuous variables are expressed as the mean and standard deviation. Categorical variables are expressed as counts (percentages). Nonparametric test, e.g., Fischer exact test whenever necessary. Statistical significance was determined at p ≤ 0.05.
Result
A total of 50 children with cystic fibrosis were included in the study. The median age of the participants was 30 months, with a range of 6 to 120 months. Most of the children were aged between > 1 and 5 years (n = 30, 60%), and the majority were male (n = 33, 66%). Among all, 57.14% of children (n = 28) had a family history of cystic fibrosis, and the parents of 41.67% of children (n = 15) had consanguinity of marriage (Table 1).
Of all, 84% of children (n = 42) yielded at least one microorganism in the culture study of respiratory specimens. Among infants (≤ 1 year), 100% of participants (n = 9) were positive for microorganisms, between > 1 and 5 years old, 80% (n = 24) were positive, and among > 5 years old, 81.8% (n = 9) were positive (Fig. 1). However, the age-related differences was not statistically significant (p = 0.477).
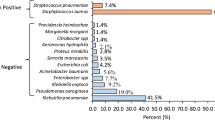
Single and double pathogens were identified in 72% (n = 36) and 12% (n = 6) of children, respectively. The most common microorganism identified from respiratory specimens was Pseudomonas aeruginosa (n = 18, 36%), followed by Klebsiella pneumoniae (n = 9, 18%), Staphylococcus aureus (n = 8, 16%), Escherichia coli (n = 8, 16%), Acinetobacter species (n = 4, 8%) and Streptococcus pneumoniae (n = 1, 2%), in decreasing order (Table 2).
Table 3 describes the antimicrobial sensitivity pattern of microorganisms identified from respiratory specimens of children with cystic fibrosis. The most sensitive drug against all strains of bacteria isolated was levofloxacin (100% sensitive against all strains), and the most resistant drugs were amoxicillin, erythromycin, and rifampicin (nearly 100% resistant against all strains). The Pseudomonas cefepime (55.56%), imipenem (50%), and meropenem (44.44%) were moderately sensitive to levofloxacin. Cefepime was 100% sensitive against Klebsiella alongside levofloxacin, followed by moderately sensitive ciprofloxacin (62.5%) and meropenem (44.44%). Staphylococcus imipenem (71.43%), vancomycin (57.14%), and ciprofloxacin (57.14%) were more sensitive than the others. Linezolid (50%), amikacin (50%), and gentamicin (50%) were moderately sensitive against Escherichia. Cotrimoxazole was the most sensitive (100%) against Acinetobacter along with levofloxacin. However, imipenem and meropenem were 75% and 50% sensitive to it, respectively. Only one case yielded Streptococcus in culture, and vancomycin, imipenem, and meropenem were 100% sensitive to Streptococcus.
A total of 16% of children (n = 8) died in the hospital, and 84% (n = 42) were discharged after improvement (Fig. 2).
Death was not significantly associated with any particular microorganism isolated from respiratory samples among children with cystic fibrosis (p > 0.05 for all) (Table 4).
Discussion
CF patients are afflicted by a vicious cycle of infection and inflammation of the airway due to decreased airway surface liquid volume and impaired mucociliary clearance [20]. A small group of bacterial species colonizes the airway in CF, causing chronic infection [9]. Understanding the prevalence and antimicrobial susceptibility pattern of these colonizing bacteria in the context of the country is important for the appropriate management of infection in CF patients. We aimed to study this pattern in the case of children with CF in Bangladesh.
Our study revealed that Pseudomonas aeruginosa is the predominant isolate from respiratory pathogens, and nearly three-quarters of children with CF were infected with a single microorganism. Among all the antimicrobial agents tested, levofloxacin was the most sensitive, and amoxicillin, erythromycin, and rifampicin were the most resistant. There was no association of outcome with species of bacteria isolated from the respiratory specimens.
We isolated P. aeruginosa and S. aureus in 36% and 16% of specimens, respectively. Our findings corroborate those of Kabir et al. [15], who conducted a large-scale clinico-epidemiologic study of CF children in Bangladesh and found a high prevalence of P. aeruginosa (27%). Nobandegani et al. [21] found a predominance of S. aureus isolates in 54% of specimens and found P. aeruginosa in 30% of specimens. Likewise, Valenza et al. [22] found a high prevalence of S. aureus (63.3%) and P. aerugisona (50%) in their participants. However, the latter two studies included both children and adults, and Santos et al. [23] previously found that P. aeruginosa and S. aureus were the most frequently found organisms in these two groups of patients. The use of antimicrobials might also affect the distribution of causative microbes in children with CF. Hauser et al. [24] noted that during the early half of the twentieth century in children with CF, when potent antistaphylococcal drugs were not available, S. aureus was the predominant isolate from respiratory pathogens and was subsequently replaced by P. aeruginosa with the development of potent antistaphylococcals.
Another noticeable observation in this study was the presence of K. pneumoniae, a gram-negative bacterium, in the respiratory specimens of our participants, which is usually associated with healthcare-acquired pneumonia and is not a typical pathogen of CF [25]. However, the genetic flexibility of K. pneumoniae enables it to evade host immunity and colonize respiratory epithelia, which might have been the reason behind such a high prevalence in our study participants.
Antimicrobial sensitivity pattern analysis in the present study showed that some of the first-generation antimicrobials, such as amoxicillin, erythromycin, and rifampicin, were nearly completely resistant against different microorganisms isolated from children with CF. Alarmingly, the emergence of carbapenem resistance was evident from our analysis, and none of the imipenem and meropenem isolates were 100% sensitive against any organisms. A northern European study [26] among cystic fibrosis patients found the development of high resistance rates of P. aeruginosa against different antimicrobials, including carbapenems. Another study in Germany noted carbapenem resistance in both mucoid and nonmucoid P. aeruginosa [22]. Frequent use of carbapenems as a broad-spectrum antibiotic in the hospital setting might lead to the development of high resistance rates to these drugs in children with CF. This is of concern, as the options of broad-spectrum antimicrobials are becoming narrower daily.
We noted high methicillin resistance among the staphylococcal isolates in our study, which conforms to the findings of previous studies [21]. Even vancomycin was found to be highly resistant (87.5%) against S. aureus. Only fluoroquinolones such as levofloxacin, ciprofloxacin, and fourth-generation cephalosporine (cefepime) were found to be effective against these strains. All beta-lactam antibiotics were resistant to E. coli. Only levofloxacin and linezolid were effective against it.
Levofloxacin was the only drug found to be 100% sensitive against all species of bacteria found in respiratory specimens of children with CF. This, although good news for clinicians, necessitates judicial and cautious use of antibiotics in cystic fibrosis patients.
The present study did not find any association between bacterial species and the outcome of cystic fibrosis patients. However, studies [24] concerning microbial species and clinical outcome in cystic fibrosis are limited by the fact that any association, if found, does not imply causation, it is impossible to find a comparison group without microbial infection, and there is lack of antimicrobial specification because of polymicrobial infection of the patients.
Excessive use of antibiotics along with lack of patient compliance leads to rapid development of resistant strains, some of which are resistant to multiple drugs. Therefore, antibiotic guidelines should be updated regularly in light of continuously updated research findings for the context of a country. Here, we found that levofloxacin is the most sensitive drug that should be preserved for the most resistant cases. Additionally, specific antibiotics should be chosen for a specific set of bacteria according to the culture and sensitivity results of respiratory specimens in children with CF.
The limitations of our study include that it was a single-center study with a small number of patients. However, the strength of the study was to present the ongoing microbial diversity and antimicrobial sensitivity pattern in children with cystic fibrosis in Bangladesh.
Conclusion
Based on our findings, we can conclude that our antibacterial treatment strategy should be targeted on P. aeruginosa with the aid of levofloxacin and other susceptible antibiotics. Surveillance of microbiological isolation and resistance patterns should be promoted regularly to improve the care and survival of CF patients.
Availability of data and materials
Data and material are available from the corresponding authors and could be shared based on reasonable request.
Abbreviations
- CBCs:
-
Complete blood counts
- CF:
-
Cystic fibrosis
- CXRs:
-
Chest X-rays
- GLP:
-
Good Laboratory Practice
- IRB:
-
Institutional Review Board
- LFT:
-
Liver function test
- MTs:
-
Mantoux tests
- RBS:
-
Random blood sugar
References
Elborn JS (2016) Cystic fibrosis. Lancet 388(10059):2519–2531
Goetz D, Ren CL (2019) Review of cystic fibrosis. Pediatr Ann 48:e154–e161
Wine JJ, Hansson GC, König P et al (2018) Progress in understanding mucus abnormalities in cystic fibrosis airways. J Cyst Fibros 17:S35–S39
Collawn JF, Matalon S (2014) CFTR and lung homeostasis. Am J Physiol - Lung Cell Mol Physiol 307:L917–L923
Cardines R, Giufre M, Pompilio A et al (2012) Haemophilus influenzae in children with cystic fibrosis : antimicrobial susceptibility, molecular epidemiology, distribution of adhesins and biofilm formation. Int J Med Microbiol 302:45–52
Wong JK, Ranganathan SC, Hart E (2013) State of the art Staphylococcus aureus in early cystic fibrosis lung disease. Pediatr Pulmonol 1159:1151–1159
Parkins MD, Floto RA (2015) Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fi brosis. J Cyst Fibros 14:293–304
Malhotra S, Hayes D, Wozniak DJ (2019) Cystic fibrosis and. Clin. Microbiol Rev 32:138–218
Cuthbertson L, Walker AW, Oliver AE et al (2020) Lung function and microbiota diversity in cystic fibrosis. Microbiome 8:1–13
Garcia-Clemente M, de la Rosa D, Máiz L et al (2020) Impact of pseudomonas aeruginosa infection on patients with chronic inflammatory airway diseases. J Clin Med 9:3800
Smyth AR, Smith SJ, Rowbotham NJ (2020) Infection prevention and control in cystic fibrosis: one size fits all The argument against. Paediatr Respir Rev 36:94–96
Lasko MJ, Huse HK, Nicolau DP et al (2021) Contemporary analysis of ETEST for antibiotic susceptibility and minimum inhibitory concentration agreement against Pseudomonas aeruginosa from patients with cystic fibrosis. Ann Clin Microbiol Antimicrob 20:1–7
Spoonhower KA, Davis PB (2016) Epidemiology of cystic fibrosis. Clin Chest Med 37:1–8
Ratjen F, Bell SC, Rowe SM, ett al (2015) Cystic fibrosis, Nature reviews. Disease primers. Nature Publishing Group; 15010.
Kabir AL, Roy S, Habib RB, et al. (2020) Cystic fibrosis diagnosed using indigenously wrapped sweating technique: first large-scale study reporting socio-demographic, clinical, and laboratory features among the children in Bangladesh a lower middle income country. Glob Pediatr Heal. 7:2333794X20967585.
Sarkar PK, Kabir AL (2018) Cystic fibrosis: a deadly disease and the vast majority are unaware of it. Bangladesh J Child Heal 42:105–107
Ferkol T, Rosenfeld M, Milla CE (2006) Cystic fibrosis pulmonary exacerbations. J Pediatr 148:259–264
Govan J (1996) Pseudomonas, Stenotrophomonas, Burkholdelia. In: Collee JG, Fraser AG, Marmion BPSA (eds) Mackie & McCartney Practical Medical Microbiology, 14th edn. Churchill Livingstone, New York, pp 413–424
Clinical & Laboratory Standards Institute (CLSI). (2017) Performance standards for antimicrobial susceptibility testing; twenty-seventh informational supplement. CLSI Document M100-S27. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. 282.
Gibson RL, Burns JL, Ramsey BW (2003) Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168:918–951
Nobandegani NM, Mahmoudi S, Pourakbari B et al (2016) Antimicrobial susceptibility of microorganisms isolated from sputum culture of patients with cystic fibrosis: Methicillin-resistant Staphylococcus aureus as a serious concern. Microb Pathog 100:201–204
Valenza G, Tappe D, Turnwald D (2008) Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros 7:123–127
Santos V, Cardoso AV, Lopes C (2017) Cystic fibrosis - comparison between patients in paediatric and adult age. Rev Port Pneumol 23:17–21
Hauser AR, Jain M, Bar-Meir M (2011) Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev 24:29–70
Riquelme SA, Ahn D, Prince A (2018) Pseudomonas aeruginosa and Klebsiella pneumoniae adaptation to innate immune clearance mechanisms in the lung. J Innate Immun 10:442–454
Mustafa MH, Chalhoub H, Denis O et al (2016) Antimicrobial susceptibility of pseudomonas aeruginosa isolated from cystic fibrosis patients in northern Europe. Antimicrob Agents Chemother 60:6735–6741
Acknowledgements
The investigator team acknowledged the ‘Pi Research Consultancy Center’ (www.pircc.org) Bangladesh for their constant and cordial support for data analysis support, and manuscript formatting.
Funding
No external fund was used to conduct the study.
Author information
Authors and Affiliations
Contributions
The conception and design: PKS, NA, MKH, and JF. Data acquisition and data collection: PKS, NA, ST, MK, JA, KAZ, TF, SSR, and MJA. Data analysis was done by JF and MKH. Interpretation of the result: PKS, NA, ST, MK, JA, KAZ, TF, SSR, MJA, MKH, and JF. Project administration: PKA, NA, ST, MJA, and TF. First draft of the manuscript: PKS, NA, MKH, and JF. Review of the draft: PKS, NA, ST, MK, JA, KAZ, TF, SSR, MJA, MKH, and JF. Final approval: PKS, NA, ST, MK, JA, KAZ, TF, SSR, MJA, MKH, and JF. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the institutional review board (IRB) of Bangladesh Institute of Child Health, Bangladesh [ethical approval no. BICH-ERC-5–2-21].
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sarkar, P.K., Akand, N., Tahura, S. et al. Antimicrobial sensitivity pattern of children with cystic fibrosis in Bangladesh: a lesson from a specialized Sishu (Children) Hospital. Egypt Pediatric Association Gaz 70, 33 (2022). https://doi.org/10.1186/s43054-022-00127-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43054-022-00127-w