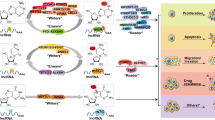
Abstract
Long noncoding RNAs are characterized as noncoding transcripts longer than 200 nucleotides in response to a variety of functions within the cells. They are involved in almost all cellular mechanisms so as epigenetics. Given that epigenetics is an important phenomenon, which participates in the biology of complex diseases, many valuable studies have been performed to demonstrate the control status of lncRNAs and epigenetics. DNA methylation and histone modifications as epigenetic mechanisms can regulate the expression of lncRNAs by affecting their coding genes. Reciprocally, the three-dimensional structure of lncRNAs could mechanistically control the activity of epigenetic-related enzymes. Dysregulation in the mutual interaction between epigenetics and lncRNAs is one of the hallmarks of cancer. These mechanisms are either directly or indirectly involved in various cancer properties such as proliferation, apoptosis, invasion, and metastasis. For instance, lncRNA HOTAIR plays a role in regulating the expression of many genes by interacting with epigenetic factors such as DNA methyltransferases and EZH2, and thus plays a role in the initiation and progression of various cancers. Conversely, the expression of this lncRNA is also controlled by epigenetic factors. Therefore, focusing on this reciprocated interaction can apply to cancer management and the identification of prognostic, diagnostic, and druggable targets. In the current review, we discuss the reciprocal relationship between lncRNAs and epigenetic mechanisms to promote or prevent cancer progression and find new potent biomarkers and targets for cancer diagnosis and therapy.
Similar content being viewed by others
Introduction
Noncoding RNAs comprise over ninety-five percent of human transcriptome [1], which are categorized into two classes based on their length, including small noncoding RNAs (including microRNAs, small interfering RNAs, and snoRNAs) and long noncoding RNAs (lncRNA) [2]. LncRNAs are the major class of noncoding RNAs whose length varies from 200 bp to 10 kb [3,4,5]. They mostly do not have any open reading frame to be translated into proteins; thus, lncRNAs have no protein-coding potential or rarely code for less than 100 amino acids [5, 6]. According to the alignment of lncRNAs with protein-coding genes, they contain introns and exons. Most of them have capping, splicing, and polyadenylation like protein-coding genes and RNAs. They are transcribed by RNA polymerase II [7] and poorly conserved with low expression levels and high tissue specificity [8,9,10]. For years, lncRNAs were known as un-functional products; however, nowadays we know them as having a wide spectrum of functions inside the cell. LncRNAs play various roles in many cellular mechanisms, such as gene expression, chromatin dynamics, apoptosis, and parental imprinting [11,12,13]. In addition, many lncRNAs can interact with microRNAs (miRs) like a sponge or compete for endogenous RNA (ceRNA) [14]. They are associated with different epigenetic phenomena and take part in two major epigenetic processes, including histone modification, through networking-related proteins such as protein members of the polycomb repressive complex (PRC2), and DNA methylation through direct and indirect cooperation with methylation-related enzymes [7, 15]. Accordingly, dysregulation in the expression of lncRNAs or their functions could affect the pathogenesis of diseases such as cancers. This could be conducted by different mechanisms, e.g., interaction with proteins or regulation of the expression of cancer-related genes [16]. Our study aims to summarize a review of how the reciprocal relationship between lncRNA and epigenetic mechanisms can promote or prevent cancer progression, and whether this field can provide new potent biomarkers and targets for cancer diagnosis and therapy.
The association of epigenetics and lncRNAs
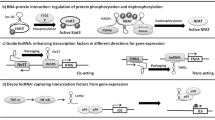
As fundamental epigenetic mechanisms such as DNA methylation and histone modifications can normally affect the expression of the protein-coding genes, they can also regulate the expression of noncoding RNAs (i.e., Figs. 1A and 2A). In this regard, some lncRNAs reciprocally can affect DNA methylation enzymes, including DNA methyltransferases (DNMTs), the ten-eleven translocation (TET), methyl-CpG-binding domains (MBDs), and histone modification enzymes. Altogether, aberrant DNA methylation and histone modification of lncRNA-coding genes or abnormal activity of DNA methylation/histone modification enzymes mediated by lncRNAs can lead to various cancers [17,18,19,20,21].
Reciprocal interaction between lncRNAs and DNA promoter methylation. A The expression level of lncRNAs is controlled by DNA methylation and DNA-methylating enzymes in the promoter region of their codding genes. B Inversely, some lncRNAs can control the expression of various genes by interacting with DNA-methylating enzymes with different mechanisms
The effect of lncRNA on different cancers
Breast cancer
Breast cancer (BC) is the leading cause of cancer deaths in women and exhibits high heterogeneity and complexity, which elevates the importance of finding novel markers for this cancer. Numerous studies have been performed on the role of lncRNAs in this cancer. For example, lncRNA MEG3 is encoded by maternally expressed gene 3, an imprinted gene that acts as a tumor suppressor gene and is dysregulated in different kinds of cancers. MEG3 regulates proliferation, migration, and invasion by inhibiting the p53 tumoral pathway. miR-506 regulates the expression of DNMT1 via SP1 and SP3 transcription factors and reduces MEG3 promoter methylation, and consequently, an increased amount of MEG3 regulates the migration and invasion of BC cells. Therefore, the downregulation of miR-506 causes hypermethylation in the MEG3 promoter and induces more proliferation, migration, and invasion in BC cells by interacting with p53 [22]. LINC00472 is another tumor suppressor regulated by promoter methylation, which is associated with better disease outcomes [1]. Additionally, Basal-like breast cancer Associated Transcript-1 (BLAT1) lncRNA is significantly upregulated in Basal-like breast cancer (BLBC) cells and this upregulation promotes the aggressive phenotype of BLBC by interfering with cell survival pathway and suppressing apoptosis [23].
Colorectal cancer
Colorectal cancer (CRC) is one of the deadliest and most highly heterogenic cancers with an increasing incidence among young adults every year [24,25,26]. CRC shows the global DNA hypomethylation of the genome and methylation in the promoter regions. According to studies on this issue, LINC00460 is overexpressed in CRC due to promoter CpGs hypomethylation. This lncRNA can be a valuable therapeutic target because it helps tumor metastasis by promoting invasion and migration [27]. LINC00460 plays an important role in CRC progression and drug resistance. This role is played through different mechanisms such as interacting with miRs such as sponging miR-939-5p or inhibition of MiR-613 [28,29,30]. PVT1, involved in EMT and angiogenesis, urothelial carcinoma-associated 1 (UCA1), and TRPM2-AS are upregulated lncRNAs in colorectal adenocarcinoma that are affected by promoter hypomethylation [31]. On the other hand, LINC00472, which is likely to be involved in cancer-related mechanisms such as cell cycle, cell migration, cell growth, and DNA repair, is downregulated in CRCs by promoter hypermethylation. Studies revealed that according to the low expression of lncRNAs, the LINC00472 promoter methylation state was a more reliable biomarker for CRC diagnosis than its expression level [32]. ZNF582-AS1 is another hypermethylated and downregulated lncRNA during CRC. This lncRNA is negatively involved in colony formation. In addition, it is hypothetically involved in G-protein coupled receptor signaling pathway [5]. Interestingly, hypermethylation in the gene body activates GATA2-AS1, which seems to correlate with poor survival in CRC [31].
Gastric cancer
Another common type of cancer with a low survival rate is GC [33], which shows a great change in epigenetic and methylation state. Previously described tumor suppressor MEG3 is significantly downregulated in GC due to hypermethylation as well. MEG3 takes part in ATP4B regulation by targeting miR-181a-5p. It also affects the expression of miR-21 so MEG3 dysregulation causes cancer promotion by increasing proliferation and apoptosis [34]. KCNKI5-ASI is another tumor suppressor that targets miR-21 and affects the levels of BCL-2, Bax, and matrix metalloproteinase (MMP)-2 and -9 [35]. Similar to other cancer types mentioned above, Linc00472 has a central role in GC by sponging miR-196a, miR-93, miR-24-3p, and several other oncomirs and suppressing cell growth and motility [36]. In addition, Linc00086 is seen to be downregulated in GC. This downregulation might be due to promoter hypermethylation under the influence of methyl-CpG-binding protein 2 (MeCP2) [37].
Cervical cancer
Cervical cancer has the 4th place of the most common and deadliest cancers among women. The vast majority of cases are caused by human papillomavirus (HPV) infection. The progress from lesions caused by HPV to cervical cancer is a long process; so early diagnosis is very important [38, 39]. LncRNA SOX21-AS1 is hypomethylated and upregulated in patients with cervical cancer. However, it acts as a tumor suppressor in many other cancers. This tumor heterogeneity gives a high prognostic value to SOX21-AS1. SOX21-AS1 is involved in sequence-specific DNA binding of RNA polymerase II transcription factor and SOX2 regulation [40]. Additionally, LINC00592 shows aberrant expression and CpG hypomethylation in cervical cancer. Its biological function has not been detected but it might have a role in transcription and cell integrity. LINC00592 also binds to two cancer-related miRs, miR-34a, and miR-449a [41]. GAS5, another methylation-effected lncRNA, is a potential biomarker and can inhibit cervical cancer. GAS5 inhibits proliferation, cell cycle progression, invasion, and migration; and additionally, induces apoptosis [42].
Pancreatic cancer
Pancreatic cancer (PC) is the 4th leading cause of cancer death worldwide. It is hard to diagnose PC in the early stages and this problem greatly reduces the chance of survival in patients. In PC cells, A2M-AS1 and DLEU2 hypomethylated in the promoter and overexpressed compared to a normal cell. MIR155HG, ITGB2-AS1, TSPOAP1-AS1, and LOC642852 are hypermethylated, but contrary to our expectation, ITGB2-AS1 shows more expression in cancer cells than healthy cells [43].
Lung cancer
Lung cancer is common cancer that has the poorest outcome among cancers [44]. NSCLC is the most common form of lung cancer with a high mortality rate. According to the studies on lncRNAs involved in NSCLC, LOC146880 positively correlates with the expression of KPNA2, a potential cancer-related gene. LOC146880 expression is associated with promoter methylation and its high expression is associated with poor overall survival in NSCLC patients [45]. In addition, AFAP1-AS1 is hypomethylated and upregulated and plays an oncogenic role in NSCLC. It can upregulate AFAP1 and promote cell migration [46].
Glioma
Glioma is the most common type of primary tumor in the brain and spinal cord that exhibits high mortality. Several lncRNAs are found to be dysregulated due to changes in promoter methylation state in glioma. Long noncoding RNA Homeobox transcript antisense intergenic RNA (HOTAIR) is one of the lncRNAs that have been studied in connection with various cancers (Table 1). HOTAIR is overexpressed in glioma and has prognostic values for both expression level and methylation state. HOTAIR expression is regulated by HOXA9 and it seems that part of HOTAIR function is mediating HOXA9 actions [47]. There are other oncogenic lncRNAs affected by hypomethylation, including H19, CYTOR, AGAAP2-AS1, MIR155HG, MIR4435-2HG, and LOC285758. The expression of these lncRNAs is associated with tumor grade and subtyping [48, 49]. Tumor suppressor MEG3 is also involved in glioma and, as mentioned above, exerts its effect through the p53 pathway [50].
Esophageal squamous cell cancer
Esophageal cancer squamous cell (ESCC) is the least studied cancer with a poor survival rate. ESCC accounts for 90% of esophageal cancer cases. Similar to other cancers described above, MEG3 plays a tumor-suppressive role in ESCC. MEG3 acts as a ceRNA for miR-9 and regulates E-cadherin and FOXO1 expression. Significant MEG3 downregulation caused by promoter hypermethylation leads to esophageal cancer progression. MEG3 similarly can downregulate mouse double minute 2 homolog (MDM2), and in this way, it can activate p53 as a cell cycle inhibitor and its target genes [51, 52]. Besides, SEMA3B-AS1, LOC100130476, and CTC-276P9.1 are other tumor suppressor lncRNAs downregulated due to hypermethylation in promoters or CpG sites nearby the transcription start site. The expression and methylation status in coding genes of these lncRNAs are associated with TNM stages, lymph node metastasis, and patients’ survival [53,54,55].
Leucemia
Myeloma
Like solid tumors, lncRNAs and the changes in their methylation states play an important role in leukemia. This association is studied much more in myeloid leukemia than in lymphoid types. For example, studies show that lncRNA H19, which is involved in BCR-ABL-mediated leukemogenesis, is overexpressed in chronic myelogenous leukemia (CML). Hypomethylation in differentially methylated regions/imprinting control regions (DMR/ICR) is responsible for H19 overexpression and accompanied by H19 expression level can be counted as a biomarker for CML [56]. HOTAIR is also found to be upregulated in CML. A higher methylation level in the HOTAIR promoter is associated with advanced CML [57]. Moreover, MEG3, which has a tumor-suppressive role by targeting miR147 and sponging miR21, shows downregulation and hypermethylation in CML [58]. In AML, MEG3 can prevent leukemogenesis both via the p53 pathway (as a cell cycle inhibitor) and in a p53-independent manner. Significant hypermethylation that is observed in the MEG3 promoter during AML occurs because of TET-2 inactivation or dysregulation in the TET2-WT1-MEG3 regulatory axis. Generally, MEG3 promoter methylation can be a prognostic marker for myeloid malignancies [4, 59, 60]. Neat1, as another tumor lncRNA, can suppress the proliferation, migration, and invasion in AML. Hypermethylation and downregulation of Neat1 promoter, affected by miR-194-5p/DNMT3A axis, lead to AML progression [61]. The BM742401 tumor suppressor is hypermethylated and downregulated in CLL [62].
Lymphoma
In the case of lymphoma and methylation-affected lncRNAs, very few studies have been performed so far. Lymphomas are classified into two categories, Hodgkin's and non-Hodgkin, but each is further subdivided into many subgroups. The most common type of lymphoma is diffuse large B-cell lymphoma (DLBCL) and is non-Hodgkin. A recent study by Zhang at el. has revealed that the tumor suppressor lncRNA NKILA is silenced duo2 to promoter hypermethylation in diffuse large B-cell lymphoma (DLBCL). NKILA is an NF-κB inhibitor so this downregulation leads to cancer progression [63]. In addition to the mentioned cases, other cancers have also been studied in this regard. For example, TMEM51-AS1 and SSTR5-AS1 in laryngeal squamous cell carcinoma, LINC00574 in bladder cancer, GAS5 in osteosarcoma, CXCL12 in thyroid carcinoma, NR_023387 in renal cell carcinoma are other instances of how methylation-related dysregulation in the expression of lncRNAs plays an important role in cancers [64,65,66,67,68,69]. Accordingly, almost in all of the deadly and prevalent types of cancer, the methylation state of promoter sites (or in some cases methylation of gene body) in lncRNA coding genes exhibits significant changes. This can be used as a diagnostic and prognostic marker and each of these genes and related proteins can be further studied as therapeutic targets.
The effect of DNA methylation on lncRNA
Many lncRNAs are detected to be significantly dysregulated because of the change in the methylation state in the promoter, or in some cases in the body of their coding gene. Aberrant DNA methylation is observed in the genome of various cancers and this can affect the biology of cancer development. Some of the lncRNAs are known for oncogenic or tumor-suppressive roles and can be used as therapeutic targets. Others can be valuable biomarkers, both for their expression level and for promoter methylation state. Therefore, lncRNAs can play a vital role in cancer development. Therefore, we have different examples of these lncRNAs in some of the most important types of cancers (Table 2).
The effect of lncRNA on epigenetics
The effect of lncRNA on DNA methylation processing
LncRNAs have a positive or negative effect on promoter DNA methylation of different genes and genome-wide methylation through various mechanisms (Fig. 1B). They can control biological processes in animals, humans, and plants. LncRNAs, similar to miRs, have oncogenic or tumor-suppressive roles in different cancers [70,71,72,73]. As a result, dysregulation of lncRNAs can lead to hyper- or hypomethylation of these genes and direct the related cells and tissues to diverse diseases, especially cancer (Table 3). Various studies have been performed on lncRNAs, which show that the increasing of lncRNAs expression affects the progression of cancer through methylation. DNA methyltransferases, as a group of the important enzymes involved in methylation, can be affected by lncRNAs. One of the most important examples is HOX Transcript Antisense RNA (HOTAIR). The upregulation of HOTAIR mechanistically correlates with hypermethylation of cyclin-dependent kinase Inhibitor 2A (CDKN2A) promoter region through miR-126 (as a negative regulator of DNMT1) repression and increasing DNMT1 expression in osteosarcoma cells [74]. In small cell lung cancer (SCLC), HOTAIR upregulation results in hypermethylation of the homeobox A1 (HOXA1) promoter region via DNMT1 and DNMT3b overexpression [75]. More examples can be mentioned in this regard. Ectopic increase of lnc-AK001058 and DACOR1 in colorectal cancer (CRC) leads to hypermethylation of A-disintegrin and metalloproteinase with thrombospondin motifs 12 (ADAMTS12) promoter and reprogramming genome-wide DNA hypermethylation of many gene promoters like JUN and FOS and consequently, the reduction of Activator protein 1 (AP-1) early response transcription factor through DNMT1, respectively [76, 77]. Studies showed that lncRNA DSCAM-AS1 was upregulated and significantly decreased miR-216b expression via its gene promoter region hypermethylation in colorectal adenocarcinoma tissues compared with marginal tissues [78]. Linc00441 is the other lncRNA that was overexpressed in GC cancerous cells and hypermethylated RB1 and repressed its expression through recruiting DNMT1 to the RB1 promoter [56]. Furthermore, LINC00337 and MIR210HG overexpression results in the hypermethylation and suppression of Tissue Inhibitor of Metalloproteinases 2 (TIMP2) and Voltage-dependent calcium channel subunit alpha2delta-2 (CACNA2D2) as tumor suppressor genes by recruiting and binding DNMT1 to their promoter region in non-small cell lung cancer (NSCLC), respectively [66, 79, 80]. A study performed on lncRNA H19 in lung adenocarcinoma (LUAD) cell lines demonstrated that the upregulation of H19 can hypermethylate the expression of a cell adhesion molecule, known as E-cadherin (CDH1), by recruiting DNMT3A and DNMT1 to the promoter region of this gene [81]. Additionally, in osteosarcoma cell lines and tissues, CDH1 can be downregulated by binding the G9a-DNMT1-Snail complex to its promoter under the effect of the overexpression of lncRNA NEAT1 [82]. Furthermore, DNMTs can be influenced by lncRNA TNRC6C-AS. Overexpression of lncRNA TNRC6C-AS1 promotes serine/threonine protein kinase 4 (STK4) methylation and downregulation of its expression via recruiting and binding DNMTs to the promoter of this gene and thus more activation of the Hippo signaling pathway in the thyroid carcinoma cells [83]. Also, LincGALH epigenetically can control Gankyrin by modifying the ubiquitination status of DNMT1, thereby deactivating PI3K/Akt/HIF1α and RhoA/ROCK signaling pathways in HCC [84]. In contrast to what was mentioned, several lncRNAs are downregulated in some cancers, which can affect the promoter region methylation of various oncogenes. For instance, downregulation of lncRNA HAGLR (also called HOXD-AS1) as a tumor suppressor lncRNA decreases DNMT1 recruiting to the promoter region of the E2F1 gene, consequently, hypomethylates this gene and increases tumor growth in LUAD tissues [85]. Furthermore, some lncRNAs affect DNA methylation with the normal route. For example, upregulation of lncRNA slincRAD in the early differentiation stages of 3T3-L1 cell hypermethylates p21 gene promoter through DNMT1 recruitment to this region in mouse adipogenesis [86]. Some lncRNAs affect DNMTs expression directly. For instance, lncRNA Rhabdomyosarcoma 2-Associated Transcript (RMST) directly increases the expression of DNMT3B by enhancing HuR as an mRNA stabilizing factor, which binds to the 3' UTR region of DNMT3B mRNA and increases its stability in the Rhabdomyosarcoma both in human and mouse. So this is one of the regulation mechanisms of the global DNA methylation level [87]. The mentioned interactions also had been known between DBCCR1‑003, lncRNA PVT1, PAUPAR lncRNA, lncRNA H19, 91H, RP5-833A20.1, ADAMTS9AS2, LINC00470, PCAT-14, ZNFX1-AS 1, lncRNA GIHCG, GAS8-AS1, and DNA methylation in bladder cancer, prostate cancer, uveal melanoma, lung cancer, esophageal squamous cell carcinoma, glioma, esophageal cancer, glioblastoma (GBM), and HCC, respectively [72, 88,89,90,91,92,93,94,95,96,97,98].
LncRNAs affecting methylation via TET family
TET family is the other DNA methylation-related enzymes that can directly or indirectly be modulated by the expression of lncRNAs and affect the DNA methylation of different genes [99]. For example, overexpression of lncRNA X-inactive specific transcript (XIST) decreases the expression of p53 mechanistically through binding to TET1 in bladder cancer [76]. The downregulation of tumor suppressor lncRNA-AC016405.3 and FENDRR as ceRNAs leads to a reduction of TET2 through miR-19a‐5p and miR-214-3P overexpression, which targets TET2 gene in the GBM cells and gastric cancer (GC) cell lines, respectively. In this way, lncRNAs can regulate the function of the TET2 enzyme [70, 100]. This association can also be seen among MAGI2-AS3 and DNA demethylation of the LRIG1 promoter mediated by TET2 in adult acute myeloid leukemia (AML) [101].
LncRNAs affecting methylation via EZH2
Several lncRNAs also have been observed with regulatory functions concerning the enhancer of zeste homolog 2 (EZH2). For instance, dysregulation of HOTAIR affects the expression of DNMTs via recruiting EZH2 to the promoter of miR-122 in hepatocellular carcinoma (HCC) [102]. Moreover, in gastrointestinal stromal cell tumors (GIST), increased expression of HOTAIR has a dual role. It can hypermethylate some CpG sites by recruiting the PRC2 to these regions and facilitating the interaction between DNMTs, EZH2, and PRC2. Simultaneously, HOTAIR can hypomethylate some other CpG dinucleotides in GIST [103]. Additionally, the upregulation of lncRNA SNHG14 augments EZH2 expression through the stabilization of its mRNA by interplaying with fused in sarcoma (FUS). It also releases EZH2 mRNA from miR-186-5p-mediated silence, thereby causing hypermethylation of the Ephrin type-A receptor 7 (EPHA7) promoter region as a developmental event mediating gene in CRC cell lines compared to normal colon tissues [104]. Moreover, the ectopic overexpression of lncRNA SNHG3 results in hypermethylation of the Mediator complex subunit 18 (Med18) gene promoter via binding to EZH2 and regulates this neighboring gene expression in GC cell lines [105]. In conclusion, lncRNAs may affect the up-/downregulation of different genes through directly or indirectly modulating DNA methylation and related enzymes, and this can lead to the progression of various cancer-related processes. Therefore, three-dimensional structures of related lncRNAs can be used as therapeutic aims to control methylation status in target cells.
Methylation of lncRNA transcripts
Another type of methylation that occurs in lncRNA transcripts can play an important role in the three-dimensional structure, stability, absorption, and binding of the lncRNA molecule. N6-methyl-adenosine (m6A) is the most abundant posttranscriptional modification in mammalian RNAs, which is carried out by a methyltransferase complex that contains at least one methyltransferase like 3 (METTL3) subunit. It is a dynamic and reversible regulation that varies in different conditions, for example, in response to cellular stress [106]. This modification is found not only in mRNAs but also in other types of RNAs such as lncRNAs [107]. The functions of m6A and the consequences of dysregulation in this modification are largely unknown because m6a cannot be detected by usual methods and does not affect the base-pairing pattern [106]. The m6A modification exerts direct control over the RNA metabolism, including mRNA processing, mRNA exporting, translation initiation, mRNA stability, and the biogenesis of lncRNA, thereby influencing various aspects of cell function. There are three main groups of proteins in association with m6A: m6A writers, erasers, and readers [108]. Dysregulation in m6a or its related proteins can lead to tumor genesis. Recent studies show that m6a lncRNA modification is associated with different types of cancer. Moreover, m6a may be involved in cancer progression by affecting lncRNA stability and accumulation. For example, lncRNA RP11 is overexpressed in CRC and promotes migration, invasion, and epithelial–mesenchymal transition (EMT). Furthermore, m6A increases RP11 nuclear accumulation and upregulates, thereby preventing Zeb1 (an EMT-related TF) degeneration [109]. Moreover, m6a stabilizes and upregulates LINC00958 in HCC. LINC00958 upregulates hepatoma-derived growth factor (HDGF) expression by sponging miR-3619-5p and helps lipogenesis and cancer progression [110]. Additionally, lncRNA GAS5 facilitates YAP translocation from the nucleus to the cytoplasm and mediates YAP degeneration so it can regulate the Hippo pathway and suppress CRC progression. Finally, m6a modification and m6A reader YTHDF3 lead GAS5 to degeneration and play an oncogenic role in CRC [111]. On the other side, lncRNAs can mediate m6a modification of other RNAs. For example, LINC00470, known as an oncogene, is overexpressed in GC tissue. LINC00470 interaction with m6A writer METTL3 and m6A reader protein YTHDF2 leads PTEN mRNA to degradation [112]. Some of the m6a-associated proteins seem to be involved in different cancers. ALKBH5, an important m6A eraser, is involved in different types of cancers by influencing different lncRNAs. Neat1 plays oncogenic roles such as sponging tumor suppressor miRs and inhibiting the expression of miR-129. In GC, overexpression of ALKBH5 upregulates NEAT1 by decreasing its m6A level. Also, ALKBH5 binds NEAT1 and affects the expression of EZH2, invasion, and metastasis [113]. Furthermore, ALKBH5 protects plasmacytoma variant translocation 1 (PVT1) against degeneration by removing m6A marks and prevents to recognize by the reader protein YTHDF2 in osteosarcoma. This overexpression of PVT1 promotes osteosarcoma cell proliferation [73]. In pancreatic cancer (PC), ALKBH5 and KCNK15-AS1 are both downregulated. lncRNA KCNK15-AS1 inhibits migration and invasion by controlling cell motility. Studies show that KCNK15-AS1 can inhibit PC by demethylating KCNK15-AS1 and may be an important therapeutic target [114]. Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) is an m6a reader protein that is demonstrated to have a role in cancer, but the exact mechanism is unknown. It sounds IGF2BP2 interacts with lncRNA DANCR via its m6a marks and together they can have a leading role in PC [76]. Long Intergenic Noncoding RNA for IGF2BP2 Stability (LINRIS) is another lncRNA interacting with IGF2BP2. LINRIS stabilizes IGF2BP2 by blocking K139 ubiquitination. The LINRIS-IGF2BP2-MYC axis has a positive role in CRC progression [115]. Accordingly, N6-methyl-adenosine and related proteins have a crucial role in cancer development. This topic has been less studied compared to other aspects of cancer epigenetics and further studies can lead us to new biomarkers and therapeutic aims.
Interaction of lncRNA and histone modification
Histone modifications like histone acetylation, and methylation, along with others, are epigenetic signatures called epigenetic codes that play vital roles in the regulation of different transcript expressions such as coding and noncoding RNAs (Fig. 2A). Reciprocally, noncoding RNAs, including lncRNAs can mechanistically regulate such histone modifications (Fig. 2B). LncRNAs have important interactions with these modifications and are known as epigenetic modulatory components that can directly or indirectly affect histone modifications and implicate various diseases. The mutual association between histone modifications and lncRNAs is proven. These interactions can lead cells to different cancers [116, 117]. There are few studies on the influence of histone modifications on lncRNAs in different cancers.
Histone acetylation/deacetylation
Tumor suppressor lncRNA BANCR is an example of lncRNAs being regulated via histone deacetylation. BANCR is downregulated in NSCLC tissue in contrast to healthy tissue. This reduction mechanistically happens through overexpression of histone deacetylase (HDAC), especially HDAC3, thereby playing a key role in EMT and metastasis in related cancers [118]. FENDRR is another tumor suppressor lncRNA that is downregulated by histone deacetylation in GC cells compared to normal samples. This downregulation leads to arise of cancerous cells toward malignancy via increasing the levels of fibronectin1 (FN1) and MMP-2/-9 gene expression [119]. Furthermore, tumor-suppressive LINC01089, which is known as lncRNA Inhibiting Metastasis (LIMT), is decreased under the influence of histone deacetylation increasing at the related promoter region in the aggressive BC patients and enhances the mammary cells’ invasion and metastasis [120]. About histone acetylation, we can mention HOXA-AS3 which is upregulated in A549 cell lines and tissue of LUAD mechanically through its unusual histone acetylation and provides a progression of cancer by forming lncRNA-HOXA6 mRNA duplex and aggregation of HOXA6 expression [121]. Besides, aberrant overexpression of LINC00152 via histone acetylation and consequently an increase of CCNE1, STAT1, STAT3, CREB1, c-MYC, and p38a proteins result in proliferation and poor survival in NSCLC [122]. On the other hand, lncRNAs can affect histone deacetylation and histone acetylation. For example, lncRNA ID2-AS1 is a tumor suppressor lncRNA that can control chromatin modification of its adjacent gene inhibitor of DNA binding 2 (ID2) via blocking HDAC8 binding to the ID2 enhancer. ID2-AS1 is downregulated in HCC cells and tissues [123]. Additionally, overexpression of lncRNA CASC9 can increase H3K27 acetylation in the LAMC2, an upstream inducer of the integrin pathway, and promote ESCC metastasis in association with other factors [124]. Concerning the association with the effect of histone methylation on lncRNA, HOXC-AS3 is a novel oncogenic lncRNA in GC cells that is significantly overexpressed due to the gain of H3K27ac and H3K4 trimethylation (H3K4me3). This abnormal histone methylation of HOXC-AS3 facilitates tumorigenesis via binding to YBX1 [125]. In breast cancer cells, UCA1 lncRNA expression is upregulated by an aberrant decrease of H3K27 trimethylation (H3K27me3) and an aberrant increase of H3K4me3 [126]. Several studies have also been done on the opposite side of this two-way interaction and the effect of lncRNAs on histone methylation. For instance, HOTAIR is a lncRNA whose effect on the various histone modifications was confirmed in some cancers. Studies show that the expression of this lncRNA increases, which regulates histone methyltransferase EZH2, histone H3K27 demethylase JMJD3, and other factors, and therefore, it leads to histone demethylation at various gene loci, e.g., SNAI1, and histone methylation in the others, including PCDH10 in renal cell carcinoma (RCC) tissues compared with non-tumoral tissues [127]. Additionally, HOTAIR can affect cyclin J (CCNJ) gene regulation and the development of cancerous cells by both binding to lysine (K)-specific demethylase 1A (LSD1) and PRC2 on different ends of these chromatin modifiers. Therefore, it causes an imbalance in miR-205 promoter H3K4me3 demethylation and H3K27 methylation and miR inhibition in bladder cancer [128]. Two studies showed that overexpression of lncRNA PVT1 could directly impact histone methylation of ANGPTL4 (angiopoietin-like 4) promoter region and indirectly downregulate EZH2 through miRNA-526b targeting at miRNA‑526b/EZH2 loop in cholangiocarcinoma (CCA) and NSCLC cells, respectively [129, 130]. Moreover, lncRNAs can influence gene histone methylation by interacting with MBD1. For instance, H19 lncRNA has an important role in histone methylation of some genes such as paternally expressed gene 1 (Peg1), insulin-like growth factor 2 (Igf2), and solute carrier family 38 member 4 (Slc38a4) through recruiting MBD1 and the other histone-modifying enzymes to these regions [131].
Multipole kinds of modifications
According to the studies, some lncRNAs are related to more than one type of histone modification in some types of cancer. For example, linc00460 lncRNA has an oncogenic role in ESCC and is overexpressed through many histone modifications, including histone H3 acetylation, H3K27Ac, H3K4Me3, and H3K4Me1 in collaboration with the binding of many transcription factors like P300, CEBPB, Jun, GATA2, Fos, etc. to its promoter region [132]. Moreover, HOTAIR plays an oncogenic role in BC in vivo and in vitro mediated by the effect of estradiol (E2) on its promoter's estrogen response elements (EREs) together with CBP/p300, histone methylases MLL3 and MLL1, histone H3K4-trimethylation, and histone acetylation. Therefore, this lncRNA can be affected by multihistone modification factors [133]. Reciprocally, PRC2 recruitment to E‑cadherin promoter, reduction in H3K27 acetylation, increase in H3K27 methylation, and changing of histone acetylation to methylation in EMT of GC are other effects of HOTAIR on the histone modification [134]. As it comes from these studies, the interaction between histone modification and lncRNAs is critical to find out how some cancers take place in different cells and tissue types without mutation in the lncRNA gene and investigate the transcriptional regulation roles of lncRNAs in a specific condition, which will improve our understanding of lncRNA genes.
Future perspective
The role of lncRNAs in different diseases has recently received special attention. There are also special databases dedicated to this topic such as the lncRNA disease database and lnc2cancer. Although many studies have been operated on how lncRNAs are involved in tumor genesis and epigenetics of cancer, there are still many unknown aspects in this field. There are other lncRNAs, which may be associated with cancer epigenetics and can be novel biomarkers. As declared above, the epigenetic state of a lncRNA coding gene can be a more reliable biomarker than the expression level of that lncRNA in some cases. In addition, better insight into mechanisms can show us new potential therapeutic targets. For example, mentioned lncRNAs or related proteins can be directly or indirectly targeted for therapy. Correspondingly, siRNAs, antisense RNAs, aptamers, and miRs can be used to suppress oncogenic lncRNAs or methylation inducer substances that can be recruited to induce methylation in promoter regions and prevent transcription of these oncogenes in the first place. On the contrary, demethylase elements can be used to upregulate tumor suppressor lncRNAs by eliminating methyl groups from their promoter. Topics such as N6 methyl adenosine are worthy of more attention. How other readers and eraser proteins interact with lncRNAs and other lncRNA undergoing m6a modification needs to be investigated. We tried to have an overview of what is done so far and what needs to be more focused on.
Conclusion
LncRNAs, despite being noncoding RNAs, can play key roles in the normal function of the cells and different diseases such as cancer. The expression levels of lncRNAs can be associated with cancer’s stage, grade, prognosis, and outcome. They can be involved in cancer progression or prevention via their interactions with different epigenetic mechanisms, including DNA methylation, histone methylation, histone acetylation, and histone deacetylation. In other words, the epigenetic regulation of lncRNAs and lncRNA-associated genes makes an indirectly epigenetic-mediated regulation of tumorigenic genes that occurred as the network in cancer progression. Given the importance of these associations presented in this review, more studies and supplementary experiments may shed the light on the cancer complexity and help find innovative approaches for the management of cancer in different stages and levels.
Availability of data and materials
Please contact the corresponding author for data requests.
Abbreviations
- ADAMTS12:
-
A-Disintegrin and metalloproteinase with thrombospondin motifs 12
- AML:
-
Adult acute myeloid leukemia
- AP-1:
-
Activator protein 1
- BC:
-
Breast cancer
- BLBC:
-
Basal-Like breast cancer
- BLAT1:
-
Basal-Like breast cancer Associated Transcript1
- CACNA2D2:
-
Calcium channel subunit alpha2delta-2
- CCA:
-
Cholangiocarcinoma
- CCNJ:
-
Cyclin J
- CDKN2A:
-
Cyclin-dependent kinase Inhibitor 2A
- ceRNA:
-
Competing for endogenous RNA
- CML:
-
Chronic myelogenous leukemia
- CRC:
-
Colorectal cancer
- DNMT:
-
DNA methyltransferase
- E2:
-
Estradiol
- EMT:
-
Epithelial–mesenchymal transition
- EPHA7:
-
Ephrin type-A receptor 7
- EREs:
-
Estrogen response elements
- ESCC:
-
Esophageal squamous cell carcinoma
- EZH2:
-
Enhancer of zeste homolog 2
- FN1:
-
Fibronectin1
- FUS:
-
Fused in sarcoma
- GBM:
-
Glioblastoma
- GC:
-
Gastric cancer
- GIST:
-
Gastrointestinal stromal cell tumors
- H3K4me3:
-
H3K4 trimethylation
- HCC:
-
Hepatocellular carcinoma
- HDAC:
-
Histone deacetylase
- HDGF:
-
Hepatoma-derived growth factor
- HOTAIR:
-
HOX Transcript Antisense RNA
- HOXA1:
-
Homeobox A1
- HPV:
-
Human papillomavirus
- ID2:
-
Inhibitor of DNA binding 2
- Igf2:
-
Insulin-like growth factor 2
- LIMT:
-
LncRNA Inhibiting Metastasis
- lncRNA:
-
Long noncoding RNAs
- LSD1:
-
Lysine (K)-specific demethylase 1A
- LUAD:
-
Lung adenocarcinoma
- MBDs:
-
Methyl-CpG-binding domains
- MDM2:
-
Mouse double minute 2 homolog
- MeCP2:
-
Methyl-CpG-binding protein 2
- Med18:
-
Mediator complex subunit 18
- MEG3:
-
Maternally expressed gene 3
- METTL3:
-
Methyltransferase like 3
- miR:
-
MicroRNA
- MMP:
-
Matrix metalloproteinase
- NSCLC:
-
Non-small cell lung cancer
- PC:
-
Pancreatic cancer
- Peg1:
-
Paternally expressed gene 1
- PRC2:
-
Polycomb repressive complex 2
- PVT1:
-
plasmacytoma variant translocation 1
- RCC:
-
Renal cell carcinoma
- RMST:
-
Rhabdomyosarcoma 2-associated transcript
- SCLC:
-
Small cell lung cancer
- Slc38a4:
-
Solute carrier family 38 member 4
- STK4:
-
Serine/threonine protein kinase 4
- TET:
-
Ten-eleven translocation
- TIMP2:
-
Tissue Inhibitor of Metalloproteinases 2
- UCA1:
-
Urothelial carcinoma-associated 1
- XIST:
-
X-inactive specific transcript
References
Shen Y, Wang Z, Loo LW, Ni Y, Jia W, Fei P et al (2015) LINC00472 expression is regulated by promoter methylation and associated with disease-free survival in patients with grade 2 breast cancer. Breast Cancer Res Treat 154(3):473–482
Liu Y, Sharma S, Watabe K (2015) Roles of lncRNA in breast cancer. Front Biosci 7:94–108
Malik B, Feng FY (2016) Long noncoding RNAs in prostate cancer: overview and clinical implications. Asian J Androl 18(4):568–574
Yao H, Duan M, Lin L, Wu C, Fu X, Wang H et al (2017) TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget 8(11):18337–18347
Kumegawa K, Maruyama R, Yamamoto E, Ashida M, Kitajima H, Tsuyada A et al (2016) A genomic screen for long noncoding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer. Sci Rep 6:26699
Cao J (2014) The functional role of long non-coding RNAs and epigenetics. Biol Procedures 16:11
Zhao Y, Sun H, Wang H (2016) Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. https://doi.org/10.1186/s13578-016-0109-3
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789
Qi P, Du X (2013) The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Modern Pathol Off J United States Canadian Academy Pathol Inc. 26(2):155–165.
Zampetaki A, Albrecht A, Steinhofel K (2018) Long non-coding RNA structure and function: is there a link? Front Physiol 9:1201
Li Z, Gao B, Hao S, Tian W, Chen Y, Wang L et al (2017) Knockdown of lncRNA-PANDAR suppresses the proliferation, cell cycle and promotes apoptosis in thyroid cancer cells. EXCLI J 16:354–362
Bhan A, Mandal SS (2015) LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochem Biophys Acta 1856(1):151–164
Saxena A, Carninci P (2011) Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. BioEssays News Rev Mol Cell Develop Biol 33(11):830–839
Tang T, Cheng Y, She Q, Jiang Y, Chen Y, Yang W et al (2018) Long non-coding RNA TUG1 sponges miR-197 to enhance cisplatin sensitivity in triple negative breast cancer. Biomed pharmacother Biomed Pharmacother 107:338–46
Hanly DJ, Esteller M, Berdasco M (2018) Interplay between long non-coding RNAs and epigenetic machinery: emerging targets in cancer? Philos Trans R Soc Lond B Biol Sci 373(1748):20170074
Yang G, Lu X, Yuan L (2014) LncRNA: a link between RNA and cancer. Biochem Biophys Acta 1839(11):1097–1109
Zare M, Bastami M, Solali S, Alivand MR (2018) Aberrant miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: diagnosis and therapeutic implications. J Cell Physiol 233(5):3729–3744
Lu C, Wei Y, Wang X, Zhang Z, Yin J, Li W et al (2020) DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer 19(1):28
Qin W, Shi Y, Zhu D, Li Y, Chen Y, Cui J et al (2020) EZH2-mediated H3K27me3 enrichment on the lncRNA MEG3 promoter regulates the growth and metastasis of glioma cells by regulating miR-21-3p. Eur Rev Med Pharmacol Sci 24(6):3204–3214
Wang Y, Chen W, Lian J, Zhang H, Yu B, Zhang M et al (2020) The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell Death Differ 27(2):695–710
Ma F, Lei Y-Y, Ding M-G, Luo L-H, Xie Y-C, Liu X-L (2020) LncRNA NEAT1 interacted with DNMT1 to regulate malignant phenotype of cancer cell and cytotoxic T cell infiltration via epigenetic inhibition of p53, cGAS, and STING in lung cancer. Front Genet 11:250
Wang XX, Guo GC, Qian XK, Dou DW, Zhang Z, Xu XD et al (2018) miR-506 attenuates methylation of lncRNA MEG3 to inhibit migration and invasion of breast cancer cell lines via targeting SP1 and SP3. Cancer Cell Int 18:171
Han YJ, Boatman SM, Zhang J, Du XC, Yeh AC, Zheng Y et al (2018) LncRNA BLAT1 is upregulated in basal-like breast cancer through epigenetic modifications. Sci Rep 8(1):15572
Liao Q, He W, Liu J, Cen Y, Luo L, Yu C et al (2015) Identification and functional annotation of lncRNA genes with hypermethylation in colorectal cancer. Gene 572(2):259–265
Mauri G, Sartore-Bianchi A, Russo AG, Marsoni S, Bardelli A, Siena S (2019) Early-onset colorectal cancer in young individuals. Mol Oncol 13(2):109–131
Bogner B (2019) Predictive markers of immunotherapy of colorectal cancer. Magy Onkol 63(3):192–195
Zhang H, Lu Y, Wu J, Feng J (2019) LINC00460 hypomethylation promotes metastasis in colorectal carcinoma. Front Genet 10:880
Wang L, Chen X, Sun X, Suo J (2020) Long noncoding RNA LINC00460 facilitates colorectal cancer progression by negatively regulating miR-613. Onco Targets Ther 13:7555–7569
Zhang Y, Liu X, Li Q, Zhang Y (2019) lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging. Cancer Manage Res 11:1779–1789
Meng X, Sun W, Yu J, Zhou Y, Gu Y, Han J et al (2020) LINC00460-miR-149-5p/miR-150-5p-Mutant p53 feedback loop promotes oxaliplatin resistance in colorectal cancer. Mol Therapy Nucleic Acids 22:1004–1015
Li Z, Tan H, Yu H, Deng Z, Zhou X, Wang M (2020) DNA methylation and gene expression profiles characterize epigenetic regulation of lncRNAs in colon adenocarcinoma. J Cell Biochem 121(3):2406–2415
Gu D, Li S, Ben S, Du M, Chu H, Zhang Z et al (2018) Circadian clock pathway genes associated with colorectal cancer risk and prognosis. Arch Toxicol 92(8):2681–2689
Slavin TP, Weitzel JN, Neuhausen SL, Schrader KA, Oliveira C, Karam R (2019) Genetics of gastric cancer: what do we know about the genetic risks? Trans Gastroenterol Hepatol 4:55
Ding L, Tian Y, Wang L, Bi M, Teng D, Hong S (2019) Hypermethylated long noncoding RNA MEG3 promotes the progression of gastric cancer. Aging 11(19):8139–8155
Zhang H, Zhang Z, Wang D (2019) Epigenetic regulation of IncRNA KCNKI5-ASI in gastric cancer. Cancer Manage Res 11:8589–8602
Tsai KW, Tsai CY, Chou NH, Wang KC, Kang CH, Li SC et al (2019) Aberrant DNA hypermethylation silenced LncRNA expression in gastric cancer. Anticancer Res 39(10):5381–5391
Yang Y, Li Y, Zhao Z, Sun R, Jiang Q, Zhao L et al (2018) DNA methylation contributes to silencing the expression of linc00086 in gastric cancer. Oncol Lett 16(2):1931–1936
Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J et al (2020) Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health 8(2):e191–e203
Sawaya GF, Smith-McCune K, Kuppermann M (2019) Cervical cancer screening: more choices in 2019. JAMA 321(20):2018–2019
Wang R, Li Y, Du P, Zhang X, Li X, Cheng G (2019) Hypomethylation of the lncRNA SOX21-AS1 has clinical prognostic value in cervical cancer. Life Sci 233:116708
Yuan LY, Qin X, Li L, Zhou J, Zhou M, Li X et al (2019) The transcriptome profiles and methylation status revealed the potential cancer-related lncRNAs in patients with cervical cancer. J Cell Physiol 234(6):9756–9763
Yang W, Xu X, Hong L, Wang Q, Huang J, Jiang L (2019) Upregulation of lncRNA GAS5 inhibits the growth and metastasis of cervical cancer cells. J Cell Physiol 234(12):23571–23580
Giulietti M, Righetti A, Principato G, Piva F (2018) LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis 39(8):1016–1025
Neal RD, Sun F, Emery JD, Callister ME (2019) Lung cancer. BMJ 365:l1725
Feng N, Ching T, Wang Y, Liu B, Lin H, Shi O et al (2016) Analysis of microarray data on gene expression and methylation to identify long non-coding RNAs in non-small cell lung cancer. Sci Rep 6:37233
Vahed SZ, Forouhandeh H, Tarhriz V, Chaparzadeh N, Hejazi MA, Jeon CO et al (2018) Halomonas tabrizica sp. nov., a novel moderately halophilic bacterium isolated from Urmia Lake in Iran. Antonie van Leeuwenhoek 111(7):1139–48
Xavier-Magalhães A, Gonçalves CS, Fogli A, Lourenço T, Pojo M, Pereira B et al (2018) The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget 9(21):15740–15756
Matjasic A, Popovic M, Matos B, Glavac D (2017) Expression of LOC285758, a potential long non-coding biomarker, is methylation-dependent and correlates with glioma malignancy grade. Radiol Oncol 51(3):331–341
Li Z, Tan H, Zhao W, Xu Y, Zhang Z, Wang M et al (2019) Integrative analysis of DNA methylation and gene expression profiles identifies MIR4435-2HG as an oncogenic lncRNA for glioma progression. Gene 715:144012
Li J, Bian EB, He XJ, Ma CC, Zong G, Wang HL et al (2016) Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. Int J Oncol 48(2):723–733
Dong Z, Zhang A, Liu S, Lu F, Guo Y, Zhang G et al (2017) Aberrant Methylation-Mediated Silencing of lncRNA MEG3 Functions as a ceRNA in Esophageal Cancer. Molecular Cancer Res MCR 15(7):800–810
Lv D, Sun R, Yu Q, Zhang X (2016) The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer. Tumour Biol J Int Soc Oncodevelop Biol Med. https://doi.org/10.1007/s13277-016-5426-y
Dong Z, Liang X, Wu X, Kang X, Guo Y, Shen S et al (2019) Promoter hypermethylation-mediated downregulation of tumor suppressor gene SEMA3B and lncRNA SEMA3B-AS1 correlates with progression and prognosis of esophageal squamous cell carcinoma. Clin Exp Metas 36(3):225–241
Guo W, Dong Z, Shi Y, Liu S, Liang J, Guo Y et al (2016) Aberrant methylation-mediated downregulation of long noncoding RNA LOC100130476 correlates with malignant progression of esophageal squamous cell carcinoma. Digest Liver Dis Off J Italian Soc Gastroenterol Italian Assoc Study Liver 48(8):961–969
Guo W, Liu S, Dong Z, Guo Y, Ding C, Shen S et al (2018) Aberrant methylation-mediated silencing of lncRNA CTC-276P9.1 is associated with malignant progression of esophageal squamous cell carcinoma. Clin Exper Meta 35(1–2):53–68
Zhou J, Shi J, Fu X, Mao B, Wang W, Li W et al (2018) Linc00441 interacts with DNMT1 to regulate RB1 gene methylation and expression in gastric cancer. Oncotarget 9(101):37471
Li Z, Luo J (2018) Epigenetic regulation of HOTAIR in advanced chronic myeloid leukemia. Cancer Manage Res 10:5349–5362
Li ZY, Yang L, Liu XJ, Wang XZ, Pan YX, Luo JM (2018) The long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid Leukemia. EBioMedicine 34:61–75
Lyu Y, Lou J, Yang Y, Feng J, Hao Y, Huang S et al (2017) Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia 31(12):2543–2551
Panahi Y, Fattahi A, Nejabati HR, Abroon S, Latifi Z, Akbarzadeh A et al (2018) DNA repair mechanisms in response to genotoxicity of warfare agent sulfur mustard. Environ Toxicol Pharmacol 58:230–236
Duan MY, Li M, Tian H, Tang G, Yang YC, Peng NC (2020) Down-regulation of lncRNA NEAT1 regulated by miR-194-5p/DNMT3A facilitates acute myeloid leukemia. Blood Cells Mol Dis 82:102417
Wang LQ, Wong KY, Li ZH, Chim CS (2016) Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget 7(50):82400–82410
Zhang MY, Calin G, Deng MD, Au-Yeung RKH, Wang LQ, Chim CS (2022) Epigenetic silencing of tumor suppressor lncRNA NKILA: implication on NF-κB signaling in non-Hodgkin's Lymphoma. Genes. 13(1)
Hui L, Wang J, Zhang J, Long J (2019) lncRNA TMEM51-AS1 and RUSC1-AS1 function as ceRNAs for induction of laryngeal squamous cell carcinoma and prediction of prognosis. PeerJ 7:e7456
Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S et al (2019) YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 38(14):2627–2644
Wu X, Li R, Song Q, Zhang C, Jia R, Han Z et al (2019) JMJD2C promotes colorectal cancer metastasis via regulating histone methylation of MALAT1 promoter and enhancing β-catenin signaling pathway. J Exper Clin Cancer Res CR 38(1):435
Xu L, Xia C, Xue B, Sheng F, Xiong J, Wang S (2018) A promoter variant of lncRNA GAS5 is functionally associated with the development of osteosarcoma. J Bone Oncol 12:23–26
Zhang S, Wang Y, Chen M, Sun L, Han J, Elena VK et al (2017) CXCL12 methylation-mediated epigenetic regulation of gene expression in papillary thyroid carcinoma. Sci Rep 7:44033
Zhou H, Guo L, Yao W, Shi R, Yu G, Xu H et al (2020) Silencing of tumor-suppressive NR_023387 in renal cell carcinoma via promoter hypermethylation and HNF4A deficiency. J Cell Physiol 235(3):2113–2128
He Z, Wang X, Huang C, Gao Y, Yang C, Zeng P et al (2018) The FENDRR/miR-214-3P/TET2 axis affects cell malignant activity via RASSF1A methylation in gastric cancer. Am J Trans Res 10(10):3211
Zhu Y, Hu S, Yao Y, Hu X (2020) LncRNA DCST1-AS1 downregulates miR-29b through methylation in glioblastoma (GBM) to promote cancer cell proliferation
Liu D, Wu K, Yang Y, Zhu D, Zhang C, Zhao S (2020) Long noncoding RNA ADAMTS9-AS2 suppresses the progression of esophageal cancer by mediating CDH3 promoter methylation. Mol Carcinog 59(1):32–44
Guo T, Gong C, Wu P, Battaglia-Hsu SF, Feng J, Liu P et al (2020) LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Diff 27(7):2191–2205
Li X, Lu H, Fan G, He M, Sun Y, Xu K et al (2017) A novel interplay between HOTAIR and DNA methylation in osteosarcoma cells indicates a new therapeutic strategy. J Cancer Res Clin Oncol 143(11):2189–2200
Fang S, Gao H, Tong Y, Yang J, Tang R, Niu Y et al (2016) Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest 96(1):60
Zheng S, Lin F, Zhang M, Fu J, Ge X, Mu N (2019) AK001058 promotes the proliferation and migration of colorectal cancer cells by regulating methylation of ADAMTS12. Am J Trans Res 11(9):5869
Somasundaram S, Forrest ME, Moinova H, Cohen A, Varadan V, LaFramboise T et al (2018) The DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA methylation in colon cancer. Clin Epigenetics 10(1):127
Zhang ZG, Zhang HS, Sun HL, Liu HY, Liu MY, Zhou Z (2019) KDM5B promotes breast cancer cell proliferation and migration via AMPK-mediated lipid metabolism reprogramming. Exp Cell Res 379(2):182–190
Yi F, Zhang P, Wang Y, Xu Y, Zhang Z, Ma W, et al. (2019) Long non-coding RNA slincRAD functions in methylation regulation during the early stage of mouse adipogenesis. RNA Biol
Kang X, Kong F, Huang K, Li L, Li Z, Wang X et al (2019) LncRNA MIR210HG promotes proliferation and invasion of non-small cell lung cancer by upregulating methylation of CACNA2D2 promoter via binding to DNMT1. Onco Targets Ther 12:3779
Gao LM, Xu SF, Zheng Y, Wang P, Zhang L, Shi SS, et al (2019) Long non‐coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation‐dependent repression of CDH1 promoter. J Cell Mol Med
Li Y, Cheng C (2018) Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am J Cancer Res 8(1):81
Peng X, Ji C, Tan L, Lin S, Zhu Y, Long M, et al (2019) Long non‐coding RNA TNRC6C‐AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway. J Cell Mol Med
Xu X, Lou Y, Tang J, Teng Y, Zhang Z, Yin Y et al (2019) The long non-coding RNA Linc-GALH promotes hepatocellular carcinoma metastasis via epigenetically regulating Gankyrin. Cell Death Dis 10(2):86
Guo X, Chen Z, Zhao L, Cheng D, Song W, Zhang X (2019) Long non-coding RNA-HAGLR suppressed tumor growth of lung adenocarcinoma through epigenetically silencing E2F1. Exp Cell Res 382(1):111461
Yi F, Zhang P, Wang Y, Xu Y, Zhang Z, Ma W et al (2019) Long non-coding RNA slincRAD functions in methylation regulation during the early stage of mouse adipogenesis. RNA Biol 16(10):1401–1413
Peng W-X, Koirala P, Zhang W, Ni C, Wang Z, Yang L et al (2019) lncRNA RMST Enhances DNMT3 Expression through Interaction with HuR. Mol Therapy. https://doi.org/10.1016/j.ymthe.2019.09.024
Ding X, Wang X, Lin M, Xing Y, Ge S, Jia R et al (2016) PAUPAR lnc RNA suppresses tumourigenesis by H3K4 demethylation in uveal melanoma. FEBS Lett 590(12):1729–1738
Fu Y, Wang W, Li X, Liu Y, Niu Y, Zhang B et al (2018) LncRNA H19 interacts with S-adenosylhomocysteine hydrolase to regulate LINE-1 Methylation in human lung-derived cells exposed to Benzo [a] pyrene. Chemosphere 207:84–90
Gao T, He B, Pan Y, Xu Y, Li R, Deng Q et al (2015) Long non-coding RNA 91H contributes to the occurrence and progression of esophageal squamous cell carcinoma by inhibiting IGF2 expression. Mol Carcinog 54(5):359–367
Kang CM, Hu YW, Nie Y, Zhao JY, Li SF, Chu S et al (2016) Long non-coding RNA RP5-833A20. 1 inhibits proliferation, metastasis and cell cycle progression by suppressing the expression of NFIA in U251 cells. Mol Med Rep 14(6):5288–96
Liu C, Fu H, Liu X, Lei Q, Zhang Y, She X et al (2018) LINC00470 coordinates the epigenetic regulation of ELFN2 to distract GBM cell autophagy. Mol Ther 26(9):2267–2281
Liu HT, Fang L, Cheng YX, Sun Q (2016) LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Cancer Med 5(12):3512–9
Qi D, Li J, Que B, Su J, Li M, Zhang C et al (2016) Long non-coding RNA DBCCR1-003 regulate the expression of DBCCR1 via DNMT1 in bladder cancer. Cancer Cell Int 16(1):81
Wang T, Ma S, Qi X, Tang X, Cui D, Wang Z et al (2016) Long noncoding RNA ZNFX1-AS1 suppresses growth of hepatocellular carcinoma cells by regulating the methylation of miR-9. Onco Targets Ther 9:5005
Wang Y, Hu Y, Wu G, Yang Y, Tang Y, Zhang W et al (2017) Long noncoding RNA PCAT-14 induces proliferation and invasion by hepatocellular carcinoma cells by inducing methylation of miR-372. Oncotarget 8(21):34429
Pan W, Zhang N, Liu W, Liu J, Zhou L, Liu Y et al (2018) The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem 293(44):17154–17165
Sui C-j, Zhou Y-m, Shen W-f, Dai B-h, Lu J-j, Zhang M-f et al (2016) Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med 94(11):1281–1296
Arab K, Karaulanov E, Musheev M, Trnka P, Schäfer A, Grummt I et al (2019) GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51(2):217–223
Ren S, Xu Y (2019) AC016405. 3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Sci 110(5):1621
Chen L, Fan X, Zhu J, Chen X, Liu Y, Zhou H (2020) LncRNA MAGI2-AS3 inhibits the self-renewal of leukaemic stem cells by promoting TET2-dependent DNA demethylation of the LRIG1 promoter in acute myeloid leukaemia. RNA Biol. 1–10
Cheng D, Deng J, Zhang B, He X, Meng Z, Li G et al (2018) LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 36:159–170
Bure I, Geer S, Knopf J, Roas M, Henze S, Ströbel P et al (2018) Long noncoding RNA HOTAIR is upregulated in an aggressive subgroup of gastrointestinal stromal tumors (GIST) and mediates the establishment of gene-specific DNA methylation patterns. Genes Chromosom Cancer 57(11):584–597
Di W, Weinan X, Xin L, Zhiwei Y, Xinyue G, Jinxue T et al (2019) Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis 10(7):1–13
Xuan Y, Wang Y (2019) Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis 10(10):1–12
Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG (2013) N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genom Proteom Bioinform 11(1):8–17
Lence T, Paolantoni C, Worpenberg L, Roignant JY (2019) Mechanistic insights into m(6)A RNA enzymes. Biochim Biophys Acta 1862(3):222–229
Chang G, Leu J-S, Ma L, Xie K, Huang S (2019) Methylation of RNA N(6)-methyladenosine in modulation of cytokine responses and tumorigenesis. Cytokine 118:35–41
Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F et al (2019) m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer 18(1):87
Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J et al (2020) M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol 13(1):5
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A et al (2019) Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer 18(1):143
Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W et al (2020) LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun 521(4):887–893
Zhang J, Guo S, Piao H-Y, Wang Y, Wu Y, Meng X-Y et al (2019) ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem 75(3):379–389
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P et al (2018) ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem 48(2):838–846
Wang Y, Lu J-H, Wu Q-N, Jin Y, Wang D-S, Chen Y-X et al (2019) LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer 18(1):174
Zhou Y, Yang H, Xia W, Cui L, Xu R, Lu H et al (2020) LncRNA MEG3 inhibits the progression of prostate cancer by facilitating H3K27 trimethylation of EN2 through binding to EZH2. J Biochem 167(3):295–301
Zhang C, Shao S, Zhang Y, Wang L, Liu J, Fang F, et al (2020) LncRNA PCAT1 promotes metastasis of endometrial carcinoma through epigenetical downregulation of E-cadherin associated with methyltransferase EZH2. Life Sci. 117295.
Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS et al (2014) Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 13(1):68
Xu TP, Xia R, Liu XX, Sun M, Yin L, Chen WM et al (2014) Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol 7(1):63
Sas-Chen A, Aure MR, Leibovich L, Carvalho S, Enuka Y, Körner C et al (2016) LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and downregulated in aggressive breast cancer. EMBO Mol Med 8(9):1052–1064
Zhang H, Liu Y, Yan L, Zhang M, Yu X, Du W et al (2018) Increased levels of the long noncoding RNA, HOXA-AS3, promote proliferation of A549 cells. Cell Death Dis 9(6):1–14
Feng S, Zhang J, Su W, Bai S, Xiao L, Chen X et al (2017) Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep 7(1):1–10
Zhou Y, Huan L, Wu Y, Bao C, Chen B, Wang L et al (2020) LncRNA ID2-AS1 suppresses tumor metastasis by activating the HDAC8/ID2 pathway in hepatocellular carcinoma. Cancer Lett 469:399–409
Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K et al (2018) LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ 25(11):1980–1995
Zhang E, He X, Zhang C, Su J, Lu X, Si X et al (2018) A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol 19(1):154
Lee J-J, Kim M, Kim H-P (2016) Epigenetic regulation of long noncoding RNA UCA1 by SATB1 in breast cancer. BMB Rep 49(10):578
Xia M, Yao L, Zhang Q, Wang F, Mei H, Guo X et al (2017) Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget 8(12):19795
Sun X, Du P, Yuan W, Du Z, Yu M, Yu X et al (2015) Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis 6(10):e1907
Qiu C, Li S, Sun D, Yang S (2020) lncRNA PVT1 accelerates progression of non-small cell lung cancer via targeting miRNA-526b/EZH2 regulatory loop. Oncol Lett 19(2):1267–1272
Yang Q, Yu Y, Sun Z, Pan Y (2018) Long non-coding RNA PVT1 promotes cell proliferation and invasion through regulating miR-133a in ovarian cancer. Biomed Pharmacother 106:61–67
Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L (2013) H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci 110(51):20693–20698
Liang Y, Wu Y, Chen X, Zhang S, Wang K, Guan X, et al (2017) A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci Rep. 37(5).
Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS (2013) Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol 425(19):3707–3722
Song Y, Wang R, Li L-W, Liu X, Wang Y-F, Wang Q-X et al (2019) Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int J Oncol 54(1):77–86
Li Z, Tan H, Yu H, Deng Z, Zhou X, Wang M (2019) DNA methylation and gene expression profiles characterize epigenetic regulation of lncRNAs in colon adenocarcinoma. J Cell Biochem
He J, Wu K, Guo C, Zhou JK, Pu W, Deng Y et al (2018) Long non-coding RNA AFAP1-AS1 plays an oncogenic role in promoting cell migration in non-small cell lung cancer. Cell mol Life Sci CMLS 75(24):4667–4681
Zhou JD, Lin J, Zhang TJ, Ma JC, Li XX, Wen XM et al (2018) Hypomethylation-mediated H19 overexpression increases the risk of disease evolution through the association with BCR-ABL transcript in chronic myeloid leukemia. J Cell Physiol 233(3):2444–2450
Di W, Weinan X, Xin L, Zhiwei Y, Xinyue G, Jinxue T et al (2019) Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis 10(7):514
Liu F, Jia J, Sun L, Yu Q, Duan H, Jiao D et al (2019) lncRNA DSCAM-AS1 downregulates miR-216b to promote the migration and invasion of colorectal adenocarcinoma cells. Onco Targets Ther 12:6789–6795
Xuan Y, Wang Y (2019) Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis 10(10):694
Zhang X, Gong J, Lu J, Chen J, Zhou Y, Li T et al (2019) Long noncoding RNA LINC00337 accelerates the non-small-cell lung cancer progression through inhibiting TIMP2 by recruiting DNMT1. Am J Transl Res 11(9):6075–6083
Gao LM, Xu SF, Zheng Y, Wang P, Zhang L, Shi SS et al (2019) Long non-coding RNA H19 is responsible for the progression of lung adenocarcinoma by mediating methylation-dependent repression of CDH1 promoter. J Cell Mol Med 23(9):6411–6428
Peng X, Ji C, Tan L, Lin S, Zhu Y, Long M et al (2020) Long non-coding RNA TNRC6C-AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway. J Cell Mol Med 24(1):304–316
Peng WX, Koirala P, Zhang W, Ni C, Wang Z, Yang L et al (2020) lncRNA RMST enhances DNMT3 expression through Interaction with HuR. Mol Ther 28(1):9–18
Hu B, Shi G, Li Q, Li W, Zhou H (2019) Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J Cell Biochem 120(4):6330–6338
Ren S, Xu Y (2019) AC016405.3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Sci 110(5):1621–32
He Z, Wang X, Huang C, Gao Y, Yang C, Zeng P et al (2018) The FENDRR/miR-214-3P/TET2 axis affects cell malignant activity via RASSF1A methylation in gastric cancer. Am J Transl Res 10(10):3211–3223
Chen L, Fan X, Zhu J, Chen X, Liu Y, Zhou H (2020) LncRNA MAGI2-AS3 inhibits the self-renewal of leukaemic stem cells by promoting TET2-dependent DNA demethylation of the LRIG1 promoter in acute myeloid leukaemia. RNA Biol 17(6):784–793
Acknowledgements
We would like to thank Dr. Behzad Baradaran for his supportive help.
Funding
This study was supported by the Immunology Research Center (grant No. 61435) of Tabriz University of Medical Sciences, Tabriz, Iran.
Author information
Authors and Affiliations
Contributions
MR and MA designed the study and drafted the manuscript. SH, MS-K, and MP were involved in data collection. ZF, FS, and DR critically revised the manuscript for important intellectual contents and MA and VT supervised the study. All listed authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ranjbar, M., Heydarzadeh, S., Shekari Khaniani, M. et al. Mutual interaction of lncRNAs and epigenetics: focusing on cancer. Egypt J Med Hum Genet 24, 21 (2023). https://doi.org/10.1186/s43042-023-00404-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-023-00404-2