Abstract
Background
Glutathione S-transferases (GSTs) are a class of important Phase II detoxification enzymes that catalyze the conjugation of glutathione and xenobiotic compounds (environmental carcinogens, pollutants and drugs) to protect against oxidative stress. GSTT1 and GSTM1 genetic polymorphisms have been extensively studied, and null genotypes or homozygous deletions have been reported in various populations. Previous studies have suggested that those who are homozygous null at the GSTM1 or GSTT1 loci are more susceptible and have a higher risk of cancers linked to environmental pollutants and drug-induced toxicity. Our study focused on GSTM1 and GSTT1 null allele frequency in the Doon population of Himachal Pradesh (India) with a comparison across other Inter and Intra-Indian ethnic groups to predict variation in the possible susceptible status.
Material and methods
Genomic DNA samples were extracted from 297 healthy unrelated individuals by a ReliaPrep™ Blood gDNA Miniprep kit (Promega, USA), and genotyped for allelic variation in GSTM1 and GSTT1 genotypes by multiplex polymerase chain reaction. Fisher's exact test was applied using SPSS.20 to analyze the genotypic distribution of GSTM1 and GSTT1 null alleles in male and female of Doon region (Solan) Himachal Pradesh.
Results
In our study, the frequency distribution of the homozygous null genotypes of GSTM1, GSTT1 individually as well as combined was found as 33.3%, 32% and 9%, respectively. Upon gender-wise comparison, a non-significant distribution (p > 0.05) for null genotypes of GSTM1 (32.8% and 35.4%, OR-0.77, 95% CI 0.42–1.41), GSTT1 (33.2% and 27.7%, OR-1.12, 95% CI 0.63–2.0) individually and combined GSTM1 and GSTT1 (10.8% and 3.7%, OR-0.31, 95% CI 0.07–1.42) were observed in studied population.
Conclusions
In our studied population, the frequency of GSTM1 null genotypes was found deviated from Inter- and Intra-Indian ethnic groups. However, the frequency of homozygous null type of GSTT1 was not significantly different, when compared to previous Indian studies, comparison with global ethnic groups showed deviation. Thus, our study has highlighted possible susceptibility risk to various xenobiotics in the Doon population of Himachal Pradesh, India.
Similar content being viewed by others
Introduction
The glutathione S-transferase (GSTs) are one of the important xenobiotics metabolizing gene family; it plays a vital role in the metabolic detoxification of oxidative stress stimulating, electrophilic compounds, carcinogens, and environmental toxins [13, 16, 37]. By their origin, GSTs are classified as cytoplasmic, mitochondrial, and membrane-associated proteins (MAPEG). Seven classes of cytoplasmic GSTs are categorized as Alpha (GSTA), Mu (GSTM), Omega (GSTO), Pi (GSTP), Sigma (GSTS), Theta (GSTT), and Zeta (GSTZ) [38].
GSTs have the ability to detoxify toxic metabolic products, like reactive nitrogen and oxygen species via glutathione peroxidase activity [9, 26]. Other than detoxification, GSTs perform a variety of biological functions including regulation of S-glutathionylation cycle, and kinase-mediated signal transduction [18, 25, 26, 41].
Previous studies have shown that deletion variations associated with GSTM1 and GSTT1 genes are located at chromosome 1 (1p13.3) and (22q11.23), respectively. Individuals having deletion variants (null/null) of GSTM1 or GSTT1 genes demonstrate a complete lack of enzymatic activity for corresponding protein [26, 28, 40]. A loss of function in the GSTs classes (M1 and T1) due to a structural deletion (Null mutation) impacts an individual’s ability to detoxify genotoxic compounds [36, 37]. GSTM1 catalyzes and detoxifies polycyclic aromatic hydrocarbon diol epoxides, such as DNA hydroperoxides, alkyl halide (component of cigarette smoke), while GSTT1 catalyzes and detoxifies benzo (a) pyrene diol epoxide, and acrolein [17, 36]. GSTM1 and GSTT1 deficiencies, either alone or in combination, are thought to have reduced detoxifying characteristics; henceforth, contribute significantly toward increased susceptibility to various types of cancer [26, 42].
In the last few decades, GSTM1 and GSTT1 null genotypes have been investigated extensively in a variety of ethnic groups, and their widespread presence has been well established [8, 11, 32, 37]. For example, the prevalence of the GSTM1 null genotype was observed 47–57% in Caucasians, 42–54% in Asians, and 16–36% in Africans, respectively, while the GSTT1 null genotype was uncommon in Caucasians (13–26%), and shown higher prevalence among Asians (35–52%) [10, 37]. These variability of GSTM1 and GSTT1 null genotypes across the worldwide population might be linked to illness susceptibilities that are unique to each population. India’s ethnic communities are diverse, both physiologically and culturally [22, 37], and northern India’s ethnic groups are particularly diverse, because of various tribal groups of Indo-European ancestry that have lived here over several waves of migration. Therefore, we have been prompted to evaluate the GSTM1, GSTT1 genotype distribution in the Northern state (Himachal Pradesh) population and to compare it with various Indian states and global populations. This is one of the first research of its type to be conducted among the people of Himachal Pradesh.
Materials and methods
Studied population and sample collection
The study was carried out among 297 healthy unrelated Himachal Pradesh residents (232 males and 65 females) with ages, ranging from 20 to 62 years. The study’s goals were well explained to all the participants and were asked to fill out a questionnaire to collect personal details as well as information related to their socioeconomic status. The research was duly approved by Institutional Ethical Committee, Maharaja Agrasen University, Baddi (HP) India.
For DNA extraction, a blood sample (5 mL) was drawn from each of the participants and collected in a vacutainer tube containing K2EDTA. All samples were transported to the laboratory in an insulated ice bucket for further processing.
Genotyping of GSTM1 and GSTT1
Genomic DNA was extracted from 200 µL of whole blood using ReliaPrep™ Blood gDNA Miniprep kit (Promega, USA). The presence or absence of the GSTM1 and GSTT1 genes was determined by multiplex-PCR. As an internal control, a portion of exon 7 of the housekeeping gene such as CYP1A1 was also co-amplified. The primer sequences [1, 20] and product size of the GSTM1 and GSTT1 genes are listed in Table 1. Both genes were genotyped using a 25-μL reaction mixture containing 1 μL of genomic DNA template (100 ng/μL), 1 μL of each primer (20 pmol/mL), 0.5 μL of dNTPs (200 μM), 2.5 μL of PCR buffer with 15 mM/L MgCl2, and 0.5 μL of Taq polymerase (3 U/μL) and 19.5 μL sterile nuclease free water. In total, 30 thermal cycles were performed. PCR reaction cycle included, initial denaturation (at 94 °C for 10 min); denaturation (at 94 °C for 60 s); annealing (at 59 °C for 45 s); and extension (at 72 °C for 60 s). The final extension was carried out at 72 °C 10 min and the amplification results were examined on 2% agarose gel.
Statistical analysis
Allelic frequencies of GSTM1 and GSTM1 were determined by direct counting and by ensuring presence or absence of DNA bands on agarose gel. Fisher’s exact test (SPSS 20; IBM, New York, USA) was applied for gender-wise distribution of both GSTs genotypes in the studied population.
Results
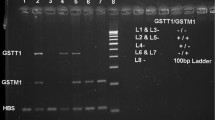
The outcomes of GSTM1 and GSTT1 genotyping, as well as the general frequency distributions of GST polymorphisms in the studied population, are shown in Fig. 1 and Table 2. In our study, the frequency distribution of the homozygous null genotypes of GSTM1, GSTT1 individually as well as combined was found as 33.3%, 32% and 9%, respectively. Upon gender-wise comparison, a non-significant distribution (p > 0.05) for null genotypes of GSTM1 (32.8% and 35.4%, OR-0.77, 95% CI 0.42–1.41), GSTT1 (33.2% and 27.7%, OR-1.12, 95% CI 0.63–2.0) individually and combined GSTM1 and GSTT1 (10.8% and 3.7%, OR-0.31, 95% CI 0.07–1.42) were observed in studied population (Table 2). Allelic distribution of GSTM1 and GSTT1 among various ethnic groups is shown in Table 3.
2% agarose gel is showing M lane is 100 pb marker, in lane 1, 3 and 7 are showing GSTT1 non-null and GSTM1 homozygous null genotyping. Lane 2 and 4 are showing GSTM1 non-null and GSTT1 homozygous null genotyping. Lane 5 is showing both GSTM1 and GSTT1 non-null genotyping and Lane 6 is showing both GSTM1 non-null and GSTT1 homozygous null genotyping
Discussion
GSTM1 and GSTT1 are the most important phase II xenobiotic metabolizing or detoxifying enzymes that catalyze glutathione conjugation, play a key role in metabolism of different types of reactive species. GST enzymes protect nucleic acids such as DNA from endogenous oxidants and save the cells by detoxifying the carcinogens and environmental contaminants. GSTs family members are major candidates for cancer disease risk, because they have the potential to regulate a person’s ability to metabolize a carcinogen. It has been reported that carries with homozygous deletions lack GST-m and GST-θ enzyme activity. The presence of deletion variations (null type) in GSTM1 and GSTT1 genes, could damage DNA and result in different types of cancer. The results of our investigation suggest that the frequencies of GSTM1 and GSTT1 null genotypes in the Doon population of Himachal Pradesh (North Indian region of Asian continent) are comparatively higher.
The allelic distribution of GSTM1 and GSTT1 among various ethnic groups has been studied (Table 3). The highest percentage of null alleles of GSTM1 (66.3%) and GSTT1 (42.5%) was observed in Asian and American populations, respectively [4, 6], while the lowest percentage of GSTM1 (11.2%) and GSTT1 (10.4%) null alleles were found in African and Asian populations, respectively [8, 27].
In our study, 33.3% of the Doon population of Himachal Pradesh had GSTM1 homozygous null or deletion genotype (Tables 2 and 3). Our results were in agreement with those of Asian, who had GSTM1 null frequencies ranging from 36.4 to 37.8% [34, 39]. Furthermore, we observed 32% of Doon population of were homozygous null or deletion for GSTT1 gene. The prevalence of homozygous null genotype of GSTT1 in in Doon population (Table 2) shows similar trend to other northern Indian populations (Table 3), [20, 31]. When compared to other Asian populations, the frequency of GSTT1 null genotype varies 10.4–27.3% which was less than and contradicting our findings [14, 27] in Asian populations. However, few studies also have shown higher percentage (35.8–40.1) of GSTT1 null genotype among different ethnic groups [8, 39]. GSTM1 and GSTT1 null frequency among Europeans varied from 45.2 to 53.5% and 16.5 to 21.3%, respectively (Table 3). GSTM1 null variants in African people varied from 11.2 to 49.5%, while GSTT1 null frequencies ranged from 22 to 35.8% (Table 3). Genotyping studies of both GSTM1 null and GSTT1 null in Asian population varied from 23.0 to 66% and 10.4 to 40.1%, respectively (Table 3). In the American, the genotype percentage of GSTM1 and GSTT1 alleles varied from 26.9 to 44.8 and 12.2 to 42.5% (Table 3). In our study, we also found non-significant differences (p > 0.05) in homozygous null genotypes of GSTM1, GSTT1 gene, individually as well as in combination among males and females in the studied population (Table 2). Upon gender-wise comparison, the non-significant differences of GSTs null genotypes might be due to the unequal number of male and female participants in the study. However, our results were in agreement with findings of a few Asians studies [10]. The difference in the gene frequency of GSTM1 and GSTT1 among the various ethnic populations is largely due to their different evolutionary histories and method selection. Our research would be useful to the Doon population of Himachal Pradesh, and globally, it will allow us to find out risk factors for disease like cancer within the susceptible individuals.
Conclusions
GSTs are involved in the detoxification of various xenobiotic compounds; therefore, knowing the prevalence of various GST alleles in a population might help in determining a risk due to susceptibility. The current study reveals the allelic distribution of GSTM1 and GSTT1 genes in the Himachal Pradesh population of the Northern region of India. Our findings have further strengthened the heterogeneity of GSTT1 and GSTM1 gene polymorphisms in the global population and also highlighted its role as a biomarker of susceptibility. From future perspectives, our findings might lay down a pathway for future bio-monitoring studies involving the risk of xenobiotic exposure in Himachal Pradesh. Moreover, our findings might add to a better understanding of the relationship between ethnicity and illness prevalence. It also might be helpful in explaining the significance of gene behavior in epidemiological and clinical research.
Availability of data and materials
The data that support the finding are available on request from corresponding author. The data are not publicly available due to privacy or ethical restriction.
References
Abdel-Rahman SM, Ahmed MMM (2007) Rapid and sensitive identification of buffalo’s, cattle’s and sheep’s milk using species-specific PCR and PCR-RFLP techniques. Food Control 18(10):1246–1249
Al-Achkar W, Azeiz G, Moassass F, Wafa AS (2014) Influence of CYP1A1, GST polymorphisms and susceptibility risk of chronic myeloid leukemia in Syrian population. Med Oncol 31:889
Al-Eitan LN, Rababa’h DM, Alghamdi MA, Khasawneh RH (2019) Association of GSTM1, GSTT1 and GSTP1 polymorphisms with breast cancer among Jordanian Women. Onco Targets Ther 12:7757–7765
Alshagga MA, Mohamed N, Suhid AN, Ibrahim IAA, Zakaria SZS (2011) Frequencies of glutathione s-transferase (GSTM1, GSTM3 and GSTT1) polymorphisms in a Malaysian population. Arch Med Sci 7:572–578
Bu H, Rosdahl I, Holmdahl-Kallen K, Sun XF, Zhang H (2007) Significance of glutathione S-transferases M1, T1 and P1 polymorphisms in Swedish melanoma patients. Oncol Rep 17:859–864
de Souza LCF, Brito TLS, Almeida AT, Muniz YP, Nishiyama PB, Souza CL, Tomazi L (2019) Association study between GSTM1 and GSTT1 genotypes and other possible risk factors in prostate cancer patients in a population from southwest bahia, brazil. Genet Mol Res 18(3):1–10
Dadbinpour A, Sheikhha MH, Darbouy M, A-Ardekani M (2013) Investigating GSTT1 and GSTM1 null genotype as the risk factor of diabetes type 2 retinopathy. J Diabetes Metab Discord 12:48
Fujihara J, Yasuda T, Iida R, Takatsuka H, Fujii Y, Takeshita H (2009) Cytochrome P450 1A1, glutathione S-transferases M1 and T1 polymorphisms in Ovambos and Mongolians. Leg Med 11:408–410
Galal AM, Walker LA, Khan IA (2015) Induction of GST and related events by dietary phytochemicals: sources, chemistry, and possible contribution to chemoprevention. Curr Top Med Chem 14:2802–2821
Garte S, Gaspar L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL (2001) Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev 10:1239–1249
Gaspar PA, Hutz MH, Salzano FM, Hill K, Hurtado AM, Petzl-Erler ML (2002) Polymorphisms of CYP1a1, CYP2e1, GSTM1, GSTT1, and TP53 genes in Amerindians. Am J Phys Anthropol 119:249–256
Gra O, Mityaeva O, Berdichevets I, Kozhekbaeva Z, Fesenko D, Kurbatova O, Goldenkova-Pavlova I, Nasedkina T (2010) Microarray-based detection of CYP1A1, CYP2C9, CYP2C19, CYP2D6, GSTT1, GSTM1, MTHFR, MTRR, NQO1, NAT2, HLA-DQA1, and AB0 allele frequencies in native Russians. Genet Test Mol Biomark 14(3):329–342
Hidaka A, Sasazuki S, Matsuo K, Ito H, Charvat H, Sawada N (2016) CYP1A1, GSTM1 and GSTT1 genetic polymorphisms and gastric cancer risk among Japanese: a nested case control study within a large-scale population-based prospective study. Int J Cancer 15:759–768
Ihsan R, Chauhan PS, Mishra AK, Singh LC, Sharma JD, Zomawia E (2014) Copy number polymorphism of glutathione-S-transferase genes (GSTM1&GSTT1) in susceptibility to lung cancer in a high-risk population from north-east India. Indian J Med Res 139:720–729
Kassogue Y, Quachouh M, Dehbi H, Quessar A, Benchekroun S, Nadifi S (2014) Effect of interaction of glutathione S-transferases (T1 and M1) on the hematologic and cytogenetic responses in chronic myeloid leukemia patients treated with imatinib. Med Oncol 31:47–54
Kasthurinaidu KP, Ramasamy T, Ayyavoo J, Dave DK, Adroja DA (2015) GST M1–T1 nullallele frequency patterns in geographically assorted human populations: a phylogenetic approach. PLoS ONE 10:e0118660
Kim WJ, Kim H, Kim CH, Lee MS, Oh BR, Lee HM, Katoh T (2002) GSTT1-null genotype is a protective factor against bladder cancer. Urol J 60:913–918
Klaus A, Zorman S, Berthier A, Polge C, Ramirez S, Michelland S, Sève M, Vertommen D, Rider M, Lentze N (2013) Glutathione S-transferases interact with AMP-activated protein kinase: Evidence for S-glutathionylation and activation in vitro. PLoS ONE 8:e62497
Klusek J, Nasierowska-Guttmejer A, Kowalik A, Wawrzycka I, Lewitowicz P, Chrapek M, Głuszek S (2018) GSTM1, GSTT1, and GSTP1 polymorphisms and colorectal cancer risk in Polish nonsmokers. Oncotarget 9(30):21224
Kumar A, Yadav A, Giri SK, Dev K, Gulati S, Gautam SK, Gupta R, Aggarwal N (2012) Allelic variation of GSTM1 and GSTT1 genes in Haryana population. Genom Med Biomark Health Sci 4(3):98–102
Lopez-Cima MF, Alvarez-Avellon SM, Pascual T, Somoano AF, Tardon A (2012) Genetic polymorphisms in CYP1A1, GSTM1, GSTP1 and GSTT1 metabolic genes and risk of lung cancer in Asturias. BMC Cancer 12:433
Majumder PP (1998) People of India: biological diversity and affinities. Evol Anthropol 6:100–110
Medjani S, Chellat-Rezgoune D, Kezai T, Chidekh M, Abadi N, Satta D (2020) Association of CYP1A1, GSTM1 and GSTT1 gene polymorphisms with risk of prostate cancer in Algerian population. Afr J Urol 26(1):1–8
Millikan R, Pittman G, Tse CK, Savitz DA, Newman B, Bell D (2000) Glutathione S-transferases M1, T1, and P1 and breast cancer. Cancer Epidemiol Biomark Prev 9(6):567–573
Pajaud J, Kumar S, Rauch C, Morel F, Aninat C (2012) Regulation of signal transduction by glutathione transferases. Int J Hepatol 2012:137676
Palma-Cano LE, Córdova EJ, Orozco L, Martínez-Hernández A, Cid M, Leal-Berumen I, Licón-Trillo A, Lechuga-Valles R, González-Ponce M, González-Rodríguez E, Moreno-Brito V (2017) GSTT1 and GSTM1 null variants in mestizo and amerindian populations from Northwestern Mexico and a literature review. Genet Mol Biol 40(4):727–735
Peddireddy V, Badabagni SP, Gundimeda SD, Mamidipudi V, Penagaluru PR, Mundluru HP (2016) Association of CYP1A1, GSTM1 and GSTT1 gene polymorphisms with risk of non-small cell lung cancer in Andhra Pradesh region of South India. Eur J Med Res 21(1):1–14
Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB (1994) Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J 300:271–276
Pinheiro DS, Rocha Filho CR, Mundim CA, Junior PM, Ulhoa CJ, Reis AA (2013) Evaluation of glutathione S-transferase GSTM1and GSTT1 deletion polymorphisms ontype-2 diabetes mellitus risk. PLoS ONE 8:e76262
Possuelo LG, Peraça CF, Eisenhardt MF, Dotto ML, Cappelletti L, Foletto E (2013) Polymorphisms of GSTM1 andGSTT1genes in breast cancer susceptibility: a case-control study. Rev Bras Ginecol Obstet 35:569–574
Ritambhara Tiwari S, Vijayraghavalu S, Kumar M (2019) Genetic polymorphisms of xenobiotic metabolizing genes (GSTM1, GSTT1, GSTP1), gene-gene interaction with association to lung cancer risk in North India; A case control study. Asian Pac J Cancer Prev 20(9):2707–2714
Saadat M (2007) GSTM1 null genotype associated with age-standardized cancer mortality rate in 45 countries from five continents: an ecologic study. Int J Cancer Res 3:74–91
Schneider J, Berger U, Philipp M, Woitowitz HJ (2004) GSTM1, GSTT1, and GSTP1 polymorphism and lung cancer risk in relation to tobacco smoking. Cancer Lett 208:65–74
Shukla RK, Tilak AR, Kumar C, Kant S, Kumar A, Mittal B (2013) Associations of CYP1A1, GSTM1 and GSTT1 polymorphisms with lung cancer susceptibility in a northern Indian population. Asian Pac J Cancer Prev 14:3345–3349
Sikdar N, Datta S, Dey B, Paul RR, Panda CK, Roy B (2005) Homozygous null genotype at glutathione S-transferase M1 locus as a risk factor for oral squamous cell carcinoma in Indian tobacco users. Int J Hum Genet 5:37–44
Strange RC, Spiter MA, Ramachandran S, Fryer AA (2001) Glutathione-S-transferase family of enzymes. Mutat Res 482:21–26
Suthar PC, Purkait P, Uttaravalli K, Sarkar BN, Ameta R, Sikdar M (2018) Glutathione S-transferase M1 and T1 null genotype frequency distribution among four tribal populations of western India. J Genet 97(1):11–24
Tew KD, Townsend DM (2012) Glutathione-s-transferases as determinants of cell survival and death. Antioxid Redox Signal 17:1728–1737
Wu W, Lu J, Tang Q, Zhang S, Yuan B, Li J (2013) GSTM1 and GSTT1 null polymorphisms and male infertility risk: an updated meta-analysis encompassing 6934 subjects. Sci Rep 3:2258
Xu S, Wang Y, Roe B, Pearson WR (1998) Characterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletion. J Biol Chem 273:3517–3527
Zhang H, Wu X, Xiao Y, Chen M, Li Z, Wei X, Tang K (2014) Genetic polymorphisms of glutathione S-transferase M1 and T1, and evaluation of oxidative stress in patients with non-small cell lung cancer. Eur J Med Res 19:67
Zhang J, Grek C, Ye ZW, Manevich Y, Tew KD, Townsend DM (2014) Pleiotropic functions of glutathione S-transferase P. Adv Cancer Res 122:143–175
Acknowledgements
Authors are duly thankful to Maharaja Agrasen University, Solan (H.P), India for providing the laboratory facility. Authors are also grateful to the persons of Himachal Pradesh who are involved in the study and provide blood sample
Funding
There was no financial assistance by any funding agency.
Author information
Authors and Affiliations
Contributions
Hemlata has done entire analysis of the research work. SKG has supervised this work. JS assists with the genotyping of genes. AB, AK and GS helped in the manuscript editing. KP has given valuable suggestions. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Participants were requested to sign a permission form (consent form), and each was questioned in person using a predetermined set of questions (standard questionnaire) which included information about their socioeconomic situation. Institutional ethical committee (IEC), Maharaja Agrasen University, Baddi (HP) India approved the study’s research methodology.
Consent for publication
All the authors have permitted the submission of the research paper in your journal.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hemlata, Singh, J., Bhardwaj, A. et al. Comparative frequency distribution of glutathione S-transferase mu (GSTM1) and theta (GSTT1) allelic forms in Himachal Pradesh population. Egypt J Med Hum Genet 23, 86 (2022). https://doi.org/10.1186/s43042-022-00298-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-022-00298-6