Abstract
Background
Homozygosity mapping is an efficient gene mapping method applicable to recessive disorders. It can detect homozygous segments of identical haplotype structures shared at a higher frequency among ventricular septal defect (VSD) and tetralogy of Fallot (TOF) cases. This study aims to identify the recessive genes involved in congenital heart disease (CHD) cases by homozygosity mapping. A total of 36 CHD cases of Indian origin were recruited based on inclusion and exclusion criteria, disease severity, and hole size. Of these, ten prediagnosed VSD and TOF cases were selected for homozygosity mapping. For in silico validation of variations, overlapping gene variants were analyzed from 26 cases based on pathogenecity and haploinsufficiency scores.
Results
Genome-wide homozygosity mapping identified 34 homozygous regions with a maximum block length of 80 bp marked for the CHD samples under study. A total of 4863 genes were identified in these 34 homozygous regions, which were present across almost all chromosomes except chromosomes 4, 8, 12, and 13. The homozygosity region found in chromosome 17 revealed genes for CHD manifestation. This homozygous region contained VSD- and TOF-related genes—Nuclear Corepressor 1 (NCOR1) and Mitogen-Activated Protein Kinase Kinase 3 (MAP2K3). In silico validation identified damaging variants for NCOR1 and MAP2K3. Three variants, G207C, C241T, and G244A, were found on exon 2 in the transcript NM_001190438 for NCOR1. Three variants were also found for MAP2K3, namely G194T and C199T in exon 5 and C578T in exon 8 in the transcript NM_002756. All these variants were present in the protein kinase domain.
Conclusion
Presence of homozygous regions identifies recessive genes leading to disease severity. Defects in recessive genes NCOR1 and MAP2K3 are responsible for abnormal myogenesis, resulting in CHD manifestation.
Similar content being viewed by others
Background
The heart is the first organ to develop in humans at around the third week of gestation. Subsequent embryonic development depends on the primordial heart’s ability to meet the embryo’s ever-increasing demand for oxygen and nutrients [1]. Heart development requires the precise migration, proliferation, and differentiation of many different cell types derived from distinct embryonic origins. These processes are controlled by signaling and molecular pathways [2]. Minor deviations from the cardiogenesis program can have severe consequences. The most common form of birth defect is congenital heart disease (CHD). They are with malformation of the heart or the large blood vessels associated with the heart and affect various parts or functions [3]. Among all the different categories of CHDs, ventricular septal defects (VSDs) are the most common defects with a prevalence rate of 2.62 per 1000 live births worldwide [4]. In the Indian population, the prevalence of ventricular septal defect (VSD) is 33%, which is the highest among all the forms of CHD [5], specifically in Mysore, 40.97% [6]. It can result from an incomplete proliferation of cardiomyocytes in the muscular septum [7].
Homozygosity mapping is an efficient gene mapping method applicable to recessive disorders. Since small chromosomal regions tend to be transmitted as whole structures, affected individuals will also have identical alleles by descent being markers located nearby the disease locus and thus will be homozygous at these markers. The method’s fundamental idea is to locate the genes involved in rare recessive traits and search for regions of homozygosity, which are shared by different affected individuals [8]. This has made it possible to identify the disease-causing mutations by performing a hypothesis-free genome-wide search for homozygosity regions through efficient homozygosity screening, using single nucleotide polymorphism (SNP) chip-based genotyping platforms [9].
In conjunction with homozygosity mapping, exome sequencing led to the identification of the causative allele for non-syndromic CHD. Homozygous haplotype mapping can be used to detect the homozygous segments of the identical haplotype structure, which is shared at a higher frequency among the VSD and tetralogy of Fallot (TOF) cases. Given this, here, we report the homozygosity haplotype region, which harbors the CHD-causing genes.
Aim
The current investigation aims to identify recessive genes involved in CHD by homozygosity mapping and pathway analysis for the identified recessive genes. These recessive genes will be validated through the overlapping study of homozygosity mapping and whole-exome sequencing for the ten samples under study.
Methods
A total of 36 cases of non-syndromic clinically prediagnosed VSD and TOF were recruited. Based on hole size and severity, seven VSD and three TOF were used for whole-exome sequencing (WES). A total of 26 cases (18 VSD and 8 TOF) were considered along with age-matched controls for validation. Karyotyping was performed to exclude trisomy cases and other chromosomal abnormalities. Echocardiogram test was conducted to calculate parameters such as hole size, misplaced aorta, hypertrophy, and valve stenosis. All subjects with a hole of radius ≥ 5 mm in the heart were considered for the study. Subjects with other CHDs malformation like atrial septal defect (ASD), aortic stenosis (AS), patent ductus arteriosus (PDA), and atrioventricular septal defect (AVSD) were excluded. Informed written consent was obtained from all the subjects, which were recruited in this study.
Genomic DNA was extracted using human peripheral blood, collected in vacutainer collection tubes using the Qiagen kit (Cat. No. 51104) following the manufacturer’s standard protocol. The isolated DNA was stored at − 20 °C for WES and validation.
Whole-exome sequencing was performed for the ten selected samples. Pair end libraries were prepared using Illumina NextSeq 2 × 75 paired-end (PE) library preparation kit from all 10 QC qualified samples. WES was performed at the coverage of 100X by using NEXTSEQ 500 machine to obtain sequence data in FASTQ format. FASTQ files consisted of four lines: (1) a sequence identifier with information about the sequencing run and the cluster—the exact contents of this line vary based on the BCL to FASTQ conversion software used; (2) the sequence (the base calls; A, C, T, G, and N); (3) a separator is simply a plus (+) sign; and (4) the base calls quality scores.
The high-quality reads were aligned against the human reference genome (hg19) for variant calls in VCF (variant calling file) format. The VCF was used as an input file for homozygosity mapping software and variant annotations. Firstly, whole-exome data (VCF) was used as an input file for homozygosity mapping. The VCF was uploaded on the homozygosity mapper software, and genotypes obtained were analyzed to identify the homozygous regions. Homozygosity mapper screened the samples for homozygous blocks using block length with a threshold of 80 bp. A homozygosity score was designated for each genotype obtained with an intuitive graphical interface for the homozygous region peaks highlighted in red [8]. These homozygous regions were visualized genome-wide with hyperlinks to directly zoom into single chromosomes and across chromosomal regions. Gene distiller was integrated within homozygosity mapper and used to map the homozygous regions to corresponding candidate genes [10]. These genes were sorted for disease correlation with the gene position in the genotype view [10]. Variant annotation was performed using variant annotator tool—wANNOVAR. This tool was based on particular algorithms with statistical significance and annotated functional consequences of genetic variants from the generated WES data. wANNOVAR output identified splicing variants and exonic variants that change protein-coding, frequency of variants observed in 1000 Genomes Project, dbSNP IDs, and mapped candidate genes for the regions.The high-quality reads were aligned against the human reference genome (hg19) for variant calls in VCF (variant calling file) format. The VCF was used as an input file for homozygosity mapping software and variant annotations. Firstly, whole-exome data (VCF) was used as an input file for homozygosity mapping. The VCF was uploaded on the homozygosity mapper software, and genotypes obtained were analyzed to identify the homozygous regions. Homozygosity mapper screened the samples for homozygous blocks using block length with a threshold of 80 bp. A homozygosity score was designated for each genotype obtained with an intuitive graphical interface for the homozygous region peaks highlighted in red [8]. These homozygous regions were visualized genome-wide with hyperlinks to directly zoom into single chromosomes and across chromosomal regions. Gene distiller was integrated within homozygosity mapper and used to map the homozygous regions to corresponding candidate genes [10]. These genes were sorted for disease correlation with the gene position in the genotype view [10]. Variant annotation was performed using variant annotator tool—wANNOVAR. This tool was based on particular algorithms with statistical significance and annotated functional consequences of genetic variants from the generated WES data. wANNOVAR output identified splicing variants and exonic variants that change protein-coding, frequency of variants observed in 1000 Genomes Project, dbSNP IDs, and mapped candidate genes for the regions.
Pathogenicity of nonsynonymous variants was predicted by sorting damaging nonsynonymous variants using pathogenicity scoring platforms (SIFT) (http://sift.bii.a-star.edu.sg/), polymorphism phenotyping V2 (PolyPhen-2) (http://genetics.bwh.harvard.edu/pph2/), and Mutation Taster (http://www.mutationtaster.org/). These damaging nonsynonymous variants were interrogated for the obtained candidate genes through homozygous mapping analysis in study subjects. The read depth (DP) of all variants considered was more than 100 to ensure the exclusion of false positives in the variant analysis. Detailed methods for WES were adapted from Gholipoorfeshkecheh et al. [11]. Variants for recessive genes obtained in homozygous regions were queried in the variant files and characterized by pathogenicity and haploinsufficiency. The data obtained from both homozygous mapping and WES was used in conjunction with the downstream analysis and silico validation.
Pathway and protein-protein interaction networks for the recessive damaging genes were constructed using the Ingenuity Pathway Analysis (IPA) tool using inbuilt curated knowledgebase for pathway and gene correlation. IPA allows searching for targeted information on genes, proteins, chemicals, and the building of interactive models of experimental systems.
Primers were designed to amplify the damaging variants using primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). The forward primer sequence was 5′ ACATGGCTCTGCTGTGGTTTA 3′, and the reverse primer sequence was 5′ GACAGTGCAGCAAGGAAAATGA 3′. Polymerase chain reaction (PCR) was performed on the thermal cycler in the total volume of 50 μl with 4 μl genomic DNA, 25 μl PCR master mix, 4 μl reverse and forward primer each, and 13 μl of distilled water with 35 amplification cycles. Each PCR cycle was performed 5 min at 95 °C, 30 s at 53 °C, and 7 min at 72 °C for 35 cycles. The PCR products were analyzed on a 1.5% agarose gel containing ethidium bromide, and 467 bp PCR product size was purified. PCR product was then subjected to Sanger sequencing.
Results
Of the 36 cases, 25 were VSD and 11 TOF cases. All these 36 cases have varied hole size defects ranging from 0.5 to 4.10 cm. Among the VSD cases, 14 cases were perimembranous, one mild muscular, eight cases subaortic, and one case outlet, while all TOF cases were subaortic. For the present study, seven VSDs and three TOFs cases were selected randomly considering the stringent characteristic features recorded in Table 1. These cases were diagnosed with a heart hole ranging from 5 to 41 mm in length. In the 2D-echocardiogram report, several episodes of dilations were observed. Three cases of VSDs showed a condition called Eisenmingerisation, which is incurable. The remaining four cases of VSDs were suitable for percutaneous device closure. Among them, only one underwent device closure through a percutaneous procedure successfully. The other three were recommended device closure. The morphology of six cases of VSD was perimembranous in nature, with one case to be sub-pulmonic. Three cases of TOF were also included for exome sequencing with an age range of 4 to 23 years (Table 1).
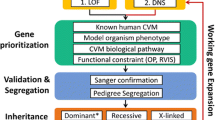
Workflow for identifying homozygous regions and candidate genes is shown in Fig. 1, which identified 34 homozygous regions within the maximum block length of 80 bp. Homozygous regions were identified in almost all chromosomes except 4, 8, 12, and 13. A total of 4863 genes were identified in the 34 homozygous regions overall. On further filtration of genes for causal CHD genes, 19 genes (NCOR1, NCOR2, NCOR3, MAP2K3, MAP2K1, MAP2K6, MAP2K7, TBX2, TBX5, TBX21, ACTN4, NKX2.1, NKX2.5, NOTCH2, HEY1, MYOCD, SMAD1, SMAD2, SMAD4) were identified to be causal for CHD. Among these homozygous regions, one of the regions present on chromosome 17 was enriched with high-confidence genes for CHD manifestation.
Recessive genes on chromosome 17, namely, NCOR1 and MAP2K3, stood out as the master genes for CHD (Fig. 2). Hence, these genes were selected for further downstream analysis. Similarly, the whole-exome sequence data analysis revealed these two genes were damaging with similar regions obtained in homozygosity mapping, resulting in CHD manifestation. The NCOR1 gene across ten subjects WES data showed three variants on exon 2—G207C, C241T, and G244A. Three damaging variants were found in MAP2K3, namely, G194T and C199T in exon 5, and C578T in exon 8 for the NM_002756 transcript. All these variants were present in the protein kinase domain (Table 2) (Fig. 3).
IPA performed in the VSD cases outlined the role of Phosphodiesterase 9A (PDE9A) in myogenesis through NCOR1 activation. Since NCOR1 protein involved deactivation of the PDE9A enzyme, the left ventricular dystrophy resulted in a series of downstream cascade steps (Fig. 4).
Pathway analysis in the VSD cases. When NCOR1 protein is affected, the product of the PDE9A enzyme does not get activated and thereby impairs the downstream cascade for PDE9A. PDE9A as a protein can no longer regulate natriuretic peptide that fails to stimulate and regulate cyclic guanosine monophosphate (cGMP) in heart myocytes. It leads to a series of complexities resulting in left ventricular dystrophy
The NCOR1 variants in the 16068377-16068377 (G207C), 16068343-16068343 (C241T), and 16068340-16068340 (G244A) regions were identified in 15.3%, 19.2%, and 46.1% of cases, respectively, while absent in controls (Table 3). Figure 5 shows PCR product of 467 bp size was obtained, and the Sanger sequence chromatogram confirmed the findings.
Discussion
Abnormalities in cardiac development are the most common form of birth defect with CHDs [12, 13]. The most important genes involved in CHDs are transcription factor genes. They are GATA4, NKX2.5, NKX2.6, T-Box, CITED2, ANKRD11, FOG2, ZIC3, and HAND2 [14]. Our laboratory’s preliminary studies revealed that two SNPs, c.608A>G and c.852G>A in the NKX2.5 gene, have an association with CHDs [15]. Earlier research from our laboratory also reported SNPs c.985 C>T in Cysteine-Rich Protein with EGF-Like Domain 1 (CRELD1) to be involved in causing CHD [16].
Echocardiography of three cases showed subaortic VSD, aortic overriding, right ventricular hypertrophy, and right ventricular outflow tract obstruction. Two TOF cases underwent intracardiac repair. The remaining one was managed medically with antibiotics because of an infected pulmonary valve, medically as infective endocarditis. The morphology of all three TOF cases was subaortic VSD in nature [17].
The homozygosity mapper tool is integrated with gene distiller to search for candidate genes and sort the candidate region’s genes. Gene distiller provided information from various data sources such as gene-phenotype associations, gene expression patterns, and protein-protein interactions [10]. Genome-wide homozygosity mapping with significant homozygous regions on chromosome 17 is marked red for both VSD and TOF cases. The presence of a homozygous region on chromosome 17 with NCOR1 and MAP2K3 genes along with its whole-exome sequence mutations aids severity towards its manifestation.
The present study identifies the presence of recessive genes NCOR1 and MAP2K3 in causing VSD and TOF through homozygosity haplotype mapping in conjunction with the WES data set where homozygous regions are identified in almost all chromosomes except 4, 8, 12, and 13. Further analysis revealed one homozygous region on chromosome 17 with genes enriched for CHD manifestation.
One of the recessive genes, NCOR1, is positioned on 17p11.2 with 54 exons with 189.31 kb length, from 16121499 to 15932191 (NCBI 37) on the reverse strand. NCOR1 is the key component of the generic corepressor complex and plays an important role in the control of heart development and differentiation [18]. The NCOR is needed for repression of myogenesis by the effect on the MYOD gene. The NCOR regulates and controls the activity of MYOD in mammalian differentiation. MYOD has a role during myogenesis, activating cell cycle, and muscle-specific gene transcription [19]. It is one of the master genes for CHD manifestation.
The other gene, MAP2K3 maps on chromosome 17 at 17q11.2, has a 30.60-kb size from 21187952 to 21218552 (NCBI 37) on the direct strand (Fig. 3). This gene is an activating transcription factor 3 (ATF3) target. ATF3 binds with the MAP2K3 promoter, recruiting histone deacetylases 1 (HDAC1), resulting in MAP2K3 gene associated histone deacetylation leading to inhibiting MAP2K3 expression. The MAP2K3 knockdown studies rescued the profibrotic/hypertrophic phenotype in ATF3KO cells. ATF3 protects against hypertensive cardiac remodeling and fibrosis, via suppressing MAP2K3 expression and p38 mitogen-activated protein kinase signaling. MAP2K3 protein is involved in P38 signaling, myogenesis, and TGF-beta signaling [17]. Hence, when the protein kinase domain in MAP2K3 is mutated, myogenesis is affected and result in CHD manifestation [20].
The physical interactors with damaging mutations affect the downstream cascade of pathways and processes, leading to disease manifestation. The NCOR1 has physically interacted with PDE9A. PDE9A as a protein regulates natriuretic peptide, stimulating and regulating cyclic guanosine monophosphate (cGMP) in heart myocytes. It is upregulated by hypertrophy and cardiac failure. When PDE9A protein expression is increased, then the cGMP-esterase activity is distinguished in left ventricular (LV) myocardium with heart failure. Therefore, the PDE9A enzyme regulates natriuretic peptide-dependent cGMP signaling in the heart, acting as a regulator of cardiac hypertrophy in myocytes [21]. Therefore, when NCOR1 protein is affected, the product of the PDE9A enzyme does not get activated. This leads to left ventricular dystrophy.
Three variants for NCOR1 present across homozygosity mapping and whole-exome sequencing are validated through Sanger sequencing. The variants of G207C and C241T are present exclusively in VSD cases. G244A variant is present in three TOF cases. These variants are not observed in any of the controls. This indicates that the variants in NCOR1 have associations with VSD and TOF phenotypes.
Limitation
The study is focused on homozygous regions in the cases. It would add to the research if parental data is also available, and homozygous regions within the family could have been identified. This will provide a holistic approach to the study, both at familial and population levels.
Conclusion
The study suggests that NCOR1 and MAP2K3 are the recessive genes, which are potentially involved in heart development. These are involved in myogenesis and mutations in these genes which result in CHD manifestation.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on a reasonable request.
Abbreviations
- ASD:
-
Atrial septal defect
- AVSD:
-
Atrioventricular septal defect
- AS:
-
Aortic stenosis
- CHD:
-
Congenital heart disease
- IHEC:
-
Institutional Human Ethics Committee
- IPA:
-
Ingenuity Pathway Analysis
- hg:
-
Human reference genome
- MAP2K3:
-
Mitogen-Activated Protein Kinase Kinase 3
- NCOR1:
-
Nuclear Corepressor 1
- PDA:
-
Patent ductus arteriosus
- PCR:
-
Polymerase chain reaction
- PDE9A:
-
Phosphodiesterase 9A
- QC:
-
Quality check
- SNP:
-
Single nucleotide polymorphism
- SNV:
-
Single nucleotide variation
- TOF:
-
Tetralogy of Fallot
- VSD:
-
Ventricular septal defect
- WES:
-
Whole-exome sequencing
References
Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL (2013) Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med 3(10):a013847
Keyte A, Hutson MR (2012) The neural crest in congenital cardiac anomalies. Differentiation 84(1):25–40
Hoffman JI (1995) Incidence of congenital heart disease: I. Postnatal incidence. Pediatr Cardiol 16(3):103–113
Menting ME, Cuypers JA, Opić P, Utens EM, Witsenburg M, van den Bosch AE et al (2015) The unnatural history of the ventricular septal defect: outcome up to 40 years after surgical closure. J Am Coll Cardiol 65(18):1941–1951
Bhardwaj R, Rai SK, Yadav AK, Lakhotia S, Agrawal D, Kumar A et al (2015) Epidemiology of congenital heart disease in India. Congenit Heart Dis 10(5):437–446
Smitha R, Karat SC, Narayanappa D, Krishnamurthy B, Prasanth SN, Ramachandra NB (2006) Prevalence of congenital heart diseases in Mysore. Indian J Hum Genet 12:11–16
Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent transcriptional response programs. Genes Dev 20(11):1405–1428
Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236(4808):1567–1570
Hildebrandt F, Heeringa SF, Rüschendorf F, Attanasio M, Nurnberg G, Becker C et al (2009) A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet 5(1):e1000353
Seelow D, Schwarz JM, Schuelke M (2008) GeneDistillerdistilling candidate genes from linkage intervals. PLoS One 3(12):e3874
Gholipoorfeshkecheh R, Agarwala S, Krishnappa S, Savitha MR, Doddaiah N, Ramachandra NB (2020) Nuclear Co-repressor 1: A Potential Candidate Gene in the Manifestation of Congenital Heart Diseases. Int J Hum Genet. 20(2):55–65
Richards A, Garg V (2010) Genetics of congenital heart disease. Curr Cardiol Rev 6(2):91–97
Okubo A, Miyoshi O, Baba K, Takagi M, Tsukamoto K, Kinoshita A et al (2004) A novel GATA4 mutation completely segregated with an atrial septal defect in a large Japanese family. J Med Genet 41(7):e97
Risebro CA, Riley PR (2006) Formation of the ventricles. Sci World J 6:1862–1880
Dinesh SM, Kusuma L, Smitha R, Savitha MR, Krishnamurthy B, Narayanappa D (2010) Single nucleotide polymorphisms of NKX2. 5 found in congenital heart disease patients of Mysore, South India. Genet Test Mol Biomarkers 14(6):873–879
Kusuma L, Dinesh SM, Savitha MR, Krishnamurthy B, Narayanappa D, Ramachandra NB (2011) A maiden report on CRELD1 single-nucleotide polymorphism association in congenital heart disease patients of Mysore, South India. Genet Test Mol Biomarkers 15(7-8):483–487
Li Y, Li Z, Zhang C, Li P, Wu Y, Wang C et al (2017) Cardiac fibroblast–specific activating transcription factor 3 protects against heart failure by suppressing MAP2K3-p38 signaling. Circulation 135(21):2041–2057
Mottis A, Mouchiroud L, Auwerx J (2013) Emerging roles of the corepressors NCOR1 and SMRT in homeostasis. Genes Dev 27(8):819–835
Bailey P, Downes M, Lau P, Harris J, Chen SL, Hamamori Y et al (1999) The nuclear receptor corepressor NCOR regulates differentiation: NCOR directly interacts with MyoD. Mol Endocrinol 13(7):1155–1168
Rose BA, Force T, Wang Y (2010) Mitogen-activated protein kinase signaling in the heart: angels versus demons in a heart-breaking tale. Physiol Rev 90(4):1507–1546
Lee DI, Zhu G, Sasaki T, Cho GS, Hamdani N, Holewinski R et al (2015) Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature 519(7544):472–488
Acknowledgements
The authors thank patients, their families, and doctors in various hospitals for samples and the University of Mysore for providing space and facilities to conduct the research. The authors also thank Dr. Azim Gholizadeh, Dr. Hadi Esfandi, Dr. Tejas Kalastavadi, Dr. Megha Murthy, Madhura U M, and lab colleagues for their support and help during the research work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
RG collected samples, performed practical works including validation, and analyzed data and manuscript writing. SA helped in conceiving the idea and designing the experiment, analyzing the whole-exome and homozygosity mapping data, network analysis, and manuscript writing. KG helped with homozygosity mapping data analysis. SK and MS diagnosed the subjects and provided the samples. NR conceived the idea, designed the experiment, and finalized the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interest
There are no competing interests in the present study.
Ethics approval and consent to participate
Prior to the start of this study, an approval from the Institutional Human Ethics Committee (IHEC-UOM No138/Ph.D/2016-17) of University of Mysore, IHEC of Mysore Medical College (MMC EC 84/16), and IHEC of JSS Medical College, Mysore (JSSMC/IEC/33/1570/2017/18), was obtained. After this, the consent was signed by the fifty members and all participants voluntarily provided their blood samples.
Consent for publication
Not applicable
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gholipoorfeshkecheh, R., Agarwala, S., G, K. et al. Whole-exome sequencing and homozygosity mapping identify variants in NCOR1 and MAP2K3 associated with non-syndromic congenital heart defects. Egypt J Med Hum Genet 21, 58 (2020). https://doi.org/10.1186/s43042-020-00101-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-020-00101-4