Abstract
Many devastating plant viruses are transmitted by insect vectors among plant hosts in a persistent-propagative manner. Pelota (Pelo) is an evolutionarily conserved protein involved in the mRNA surveillance system. In this study, it was found that the accumulation of Pelo proteins are slightly decreased during the propagation of the fijivirus southern rice black-streaked dwarf virus (SRBSDV) in rice and transmission vector planthopper (Sogatella furcifera). The tubular protein P7-1 encoded by SRBSDV interacted with Pelo of rice or planthopper vector. Overexpression or knockdown of Pelo expression inhibits the formation of P7-1 tubules in insect cells, further exerting antiviral activity. Furthermore, overexpression or knockout of Pelo expression in transgenic rice plants also inhibits the effective propagation of SRBSDV as well as two other rice viruses of different families. The slight reduction of Pelo accumulation during SRBSDV propagation in rice and insect vectors would avoid Pelo-mediated excessive inhibition of P7-1 tubule formation, ensuring effective virus propagation. Our findings provide insights into how the up- or down-regulated expression of Pelo in rice hosts and insect vectors elevate their resistance to rice viruses.
Similar content being viewed by others
Background
Translation is an important process in which messenger RNA (mRNA) is transformed into protein (Bicknell and Ricci 2017). Eukaryotes have developed at least three mRNA surveillance mechanisms which recognize and degrade defective mRNA (Passos et al. 2009; van Hoof and Wagner 2011; Simms et al. 2017). These mechanisms include no-go mRNA decay (NGD), nonsense-mediated decay (NMD), and no-stop decay (NSD). NGD degrades mRNA that is abnormally stalled, while NMD removes aberrant mRNA that contains a premature termination codon due to a base mutation, and NSD degrades mRNA that lacks a stop codon (Jamar et al. 2018; Kong et al. 2021). Meanwhile, RNA quality control (RQC) is also one of the major RNA-mediated plant immunity degradation pathways that target and degrade viral RNAs in plants (Li and Wang 2019). The well-studied RQC mechanism NMD has emerged as a contributor to viral restriction in both animals and plants (Li and Wang 2019; Zhao et al. 2020). The evolutionarily conserved protein Pelota (Pelo) can form a heterodimeric complex with Hbs1, a GTPase Hsp70 subfamily B suppressor 1, which proceeds the process of NGD (Lee et al. 2007; Tsuboi et al. 2012). Dom34, a Pelo homolog in yeast, was reported to recognize and eliminate the exogenous RNA in antiviral defense via a mechanism that in the degradation of aberrant transcripts in RNA quality control and Hbs1 could degrade HBV X-mRNA at the RNA exosome (Aly et al. 2016; Nogimori et al. 2019). In recent years, there have been reports that the Pelo genes confer virus resistance in plants and insects. For example, tomato TY172 line carrying ty-5 which encodes Pelo with an amino acid mutation has broad-spectrum resistance to geminiviruses (Lapidot et al. 2015; Wang et al. 2018; Ren et al. 2022). A recessive gene pepy-1 contains a single nucleotide polymorphism (A to G) located at the splice site of the 9th intron of CaPelota, resulting in the addition of 28 amino acids to CaPelo that confers resistance to begomoviruses (Koeda et al. 2021). SUMOylation-modified Pelo-Hbs1 RNA surveillance complex restricts the infection of potyviruses in Nicotiana benthamiana (Ge et al. 2023). The Pelo deletion of Drosophila mutant can inhibit the replication of Drosophila C virus by limiting its capsid protein synthesis, and Pelo-deficient Drosophila also resists the replication of cricket paralysis virus, Drosophila X virus, and invertebrate iridescent virus 6 (Wu et al. 2014). In addition, mutant variants of Pelo in rice exhibits disease resistance against bacterial leaf blight by activating the salicylic acid metabolic pathway (Ding et al. 2018; Zhang et al. 2018). However, little is known about the function of Pelo in restricting rice viruses in rice plants or insect vectors.
Over the past two decades, the fijivirus southern rice black-streaked dwarf virus (SRBSDV) transmitted by white-backed planthopper Sogatella furcifera has led to epidemic outbreaks and significant losses in rice yield in Southern China and Southeast Asia (Zhou et al. 2008, 2013). Meanwhile, the phytoreovirus rice dwarf virus (RDV) transmitted by rice leafhopper Nephotettix cincticeps, and the newly discovered cytorhabdovirus rice stripe mosaic virus (RSMV) transmitted by rice leafhopper Recilia dorsalis have caused serious yield losses in Southern China (Chen et al. 2012; Yang et al. 2017, 2018). SRBSDV was first discovered in Guangdong province and has spread to at least 16 provinces in China (Yin et al. 2011; Zhou et al. 2021). The virion of SRBSDV is a non-enveloped icosahedron of 70–75 nm in diameter, and its genome is consisted of 10 double-stranded RNA (dsRNA) segments which encode six putative structural proteins (P1, P2, P3, P4, P8, and P10) and seven putative nonstructural proteins (P5-1, P5-2, P6, P7-1, P7-2, P9-1, and P9-2) (Li et al. 2018). The nonstructural protein P7-1 of SRBSDV can form tubular structures that facilitate viral particles to overcome the insect midgut barrier (Liu et al. 2011; Jia et al. 2014). During the formation of P7-1 tubular structures, endoplasmic reticulum (ER) membrane proteins, such as B-cell receptor associated protein 31, DNAJB11, and DNAJB12-Hsc70 chaperone complex, are hijacked to facilitate the assembly and ER-to-cytosol transport (Yu et al. 2021; Liang et al. 2022). Besides, P7-1 induces BCL2 interacting protein 3 (BNIP3)-mediated mitophagy by promoting the formation of phosphorylated BNIP3 dimers on the mitochondria, while P10 interacts with lysosomal-associated membrane protein 1 (LAMP1) for inhibiting fusion of autophagosomes and lysosomes to facilitate persistent viral infection in insect vectors (Liang et al. 2023; Zhang et al. 2023). RDV, as the first discovered transovarially transmitted plant virus, also exploits virus-containing tubules composed of the nonstructural viral protein Pns10 to traffic along actin-based cellular protrusions, facilitating the intercellular viral spread within insect vector (Chen et al. 2012). RDV also directly exploits insect symbiotic bacterium through virus-bacterium interaction for its transovarial transmission (Jia et al. 2017). Recently, RSMV and rice gall dwarf virus (RGDV) frequently co-infected rice plants and insect vectors in the field (Jia et al. 2022). In co-infected vectors, RSMV exploits RGDV-induced autophagosomes to assemble enveloped virions to facilitate its own propagation (Jia et al. 2023). In rice, RSMV-encoded glycoprotein induces and harnesses host antiviral autophagy for maintaining its compatible infection (Huang et al. 2023). Due to the lack of germplasm resources with effective virus resistance, it is urgent to explore resistance genes in rice and insects for the prevention and control of rice viruses.
In this study, we found that the accumulation levels of Pelo proteins are slightly decreased in SRBSDV-infected rice and S. furcifera, and SRBSDV P7-1 could interact with Pelo proteins from rice and S. furcifera. However, overexpression or knockdown of Pelo expression inhibited the formation of P7-1 tubules in insect vectors. Furthermore, overexpression or knockout of Pelo expression in transgenic rice plants also inhibited the propagation of SRBSDV as well as RDV and RSMV. Our findings provide insights into how the up-regulation and down-regulation of Pelo expression in rice and S. furcifera show the resistance to effective SRBSDV propagation. The slight reduction of Pelo accumulation during vial propagation in plant hosts and insect vectors would avoid Pelo-mediated excessive inhibition of P7-1 expression, ensuring effective virus propagation.
Results
Pelo expression is slightly reduced in rice plants or insect vectors infected by SRBSDV
The full-length Pelo gene from S. furcifera (SfPelo) contained a 1185-nt ORF encoding a protein of 394 aa that includes three similar conserved domains, eRF1-1 (aa 1–130), eRF1-2 (aa 136–268), and eRF1-3 (aa 271–370) (Fig. 1a). Sequence alignment indicated that SfPelo shared about 50% amino acid similarity with Pelo protein of 378 aa from Oryza sativa (OsPelo), which is encoded by a 1137-nt ORF, and includes three similar conserved domains (Fig. 1a). Phylogenetic tree showed the amino acid sequence of Pelo from S. furcifera is highly homologous to those of N. cincticeps, R. dorsalis, and other insect species, while the amino acid sequence of Pelo from rice is highly conserved with that of other plant species (Fig. 1b). We then investigated whether Pelo was involved in SRBSDV infection. RT-qPCR and western blot assays were used to detect the relative transcript and protein accumulation levels of Pelo in nonviruliferous or viruliferous S. furcifera. The results showed that the relative transcript level of SfPelo in SRBSDV-infected insects was 79.5% of that in nonviruliferous insects (Fig. 1c). The protein accumulation level of SfPelo in SRBSDV-infected insects was 71% of that in nonviruliferous insects (Fig. 1d). In addition, both the relative transcript and protein accumulation levels of OsPelo in SRBSDV-infected rice plants were around 80% of that in uninfected rice plants (Fig. 1e, f). Thus, SRBSDV propagation leads to a slight decrease of Pelo expression in both vector insects and host plants.
SRBSDV infection slightly decreases Pelo expression in rice plants or insect vectors. a Amino acid sequence alignment of the Pelo homology from O. sativa (OsPelo) and S. furcifera (SfPelo). The alignment was performed using the software DNAMAN. Black shading indicates conserved residues. b Phylogenetic relationship of Pelo from S. furcifera, N. cincticeps, R. dorsalis, rice, and other insects and plants. Phylogenetic tree was constructed using the Neighbor-Joining method. The branch lengths are proportional to the number of nucleotide changes, as indicated by the scale bar (0.05 substitutions per site). c, d The relative transcript levels of SfPelo and protein accumulation levels of SfPelo and SRBSDV P10 in nonviruliferous and viruliferous S. furcifera, as determined by RT-qPCR and western blot assays. V−, nonviruliferous; V+, viruliferous. Means (± SD) from three biological replicates are shown. *, P < 0.05. Insect H3 served as a reference protein. The relative intensities of bands of SfPelo were determined using ImageJ. e, f The relative transcript levels of OsPelo and protein accumulation levels of OsPelo and SRBSDV P10 in healthy and SRBSDV-infected rice, as determined by RT-qPCR and western blot assays. Means (± SD) from three biological replicates are shown. *, P < 0.05. Coomassieblue-stained gel for rubisco demonstrated the loading amounts of the proteins. The relative intensities of bands of OsPelo were determined using ImageJ
SRBSDV P7-1 interacts with Pelo proteins from S. furcifera and rice plant
To identify whether Pelo interacted directly with viral proteins, yeast two-hybrid (Y2H) assay was used to investigate the interactions of viral proteins with SfPelo or OsPelo. The results showed that only nonstructural protein P7-1 directly interacted with SfPelo and OsPelo (Fig. 2a). GST pull-down assay further revealed that both SfPelo and OsPelo interacted with P7-1 of SRBSDV (Fig. 2b, c). It is known that Pelo-Hbs1 complex is conserved in different species and plays essential role in mRNA quality control (Ge et al. 2023). The interaction of Hbs1 with Pelo or P7-1 was investigated using Y2H assays. The results showed that SfHbs1 directly interacted with SfPelo, and OsHbs1 interacted with OsPelo, while the interaction between P7-1 and SfHbs1 or OsHbs1 was absent (Fig. 2d). These results suggest that the interaction between P7-1 and SfPelo or OsPelo may play a crucial role during viral infection in planthopper vectors or rice plants.
The interaction between SRBSDV P7-1 and Pelo from S. furcifera or rice. a Interaction between P7-1 and SfPelo or OsPelo in Y2H assays. Transformants were plated on DDO and QDO medium. b, c Interaction between P7-1 and OsPelo or SfPelo in GST pull-down assay. P7-1-GST was incubated with glutathione-Sepharose beads. OsPelo-His or SfPelo-His was then added to the beads, followed by western blot assay to detect Pelo bound to P7-1-GST. d Interactions between Hbs1 and Pelo of rice or S. furcifera, P7-1 and Hbs1 of rice or S. furcifera were identified by Y2H assay. Transformants were plated onto DDO and QDO. pGADT7-T + pGBKT7-P53 as positive control, pGADT7-T + pGBKT7-Lam as negative control. DDO, SD/-Trp-Leu medium. QDO, SD/-Trp-Leu-His-Ade medium
Overexpression of SfPelo inhibits the formation of P7-1 tubular structures
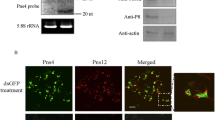
SRBSDV exploits virus-associated tubular structures, which is composed of the nonstructural membrane protein P7-1, to spread throughout the body of S. furcifera (Jia et al. 2014). We then used the recombinant baculovirus expression system to investigate the effect of overexpression SfPelo on P7-1 formation in Sf9 cells. Immunofluorescence microscopy showed that P7-1 was diffused in the cytoplasm at 24 h post-infection (hpi) in Sf9 cells co-expressed GFP or SfPelo (Fig. 3a). At 48 hpi, P7-1 formed tubular structures in the cytoplasm of Sf9 cells co-expressed GFP, but P7-1 was still diffusely distributed throughout the cytoplasm of Sf9 cells co-expressed SfPelo (Fig. 3b). At 72 hpi, P7-1 formed tubular structures in Sf9 cells co-expressed GFP or SfPelo (Fig. 3c). To further determine the effect of SfPelo expression on P7-1 accumulation, we examined the accumulation levels of the P7-1 protein. Western blot assay showed that the accumulation levels of P7-1 in Sf9 cells co-expressed SfPelo and P7-1 were lower than in that in Sf9 cells co-expressed GFP and P7-1 at 24, 48, or 72 hpi (Fig. 3d). Potentially, SfPelo suppresses the formation of P7-1 tubular structures, thereby inhibits SRBSDV propagation in insect vectors. To address this hypothesis, the prokaryotically expressed SfPelo protein was purified and microinjected into SRBSDV-infected S. furcifera to determine its role in propagation of SRBSDV in vivo. Western blot assay showed that the accumulation levels of P7-1 in SRBSDV-infected S. furcifera microinjected with SfPelo were lower than the GFP control (Fig. 3e). Taken together, our results reveal that SfPelo suppresses the propagation of SRBSDV in insect vectors.
Overexpression of SfPelo inhibits the assembly of P7-1 tubular stuctures in Sf9 cells. a–c Sf9 cells co-expressed with P7-1 and SfPelo-His were immunolabelled with P7-1-rhodamine (red) and His-FITC (green) at 24, 48, and 72 hpi, respectively. P7-1 co-infected with GFP were served as the control. Scale bars: 10 μm. d The protein accumulation levels of P7-1 in Sf9 cells co-expressed with P7-1 and GFP or SfPelo for 24, 48, and 72 h, as determined by western blot assays. GAPDH was served as reference protein. The relative intensities of bands of P7-1 protein were determined using ImageJ. e The protein accumulation levels of P7-1 in S. furcifera microinjected with SfPelo or GFP for 4 days, as determined by western blot assays. H3 was served as reference protein. The relative intensities of bands of different proteins were determined using ImageJ
Knockdown of SfPelo expression inhibits SRBSDV propagation in S. furcifera
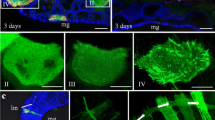
At 6 days post-first access of insects to diseased plants (padp), immunofluorescence microscopy showed the colocalization of SfPelo with P7-1-specific structures in virus-infected midguts of S. furcifera (Fig. 4a). Immunoelectron microscopy further confirmed that SfPelo antibody specifically reacted with virus-associated tubular structures in virus-infected midguts (Fig. 4b). Thus, Pelo is recruited to P7-1 tubules via interaction with P7-1. It has been reported that the knockout or mutant variants of Pelo plays a role in antiviral function (Wang et al. 2018; Koeda et al. 2021). To further determine the role of SfPelo in insect vectors during viral infection, at 6 days padp, the fifth instar nymphs of viruliferous S. furcifera were microinjected with the synthesized dsRNAs targeting SfPelo (dsSfPelo) or GFP (dsGFP). RT-qPCR and western blot assays showed that the transcript and accumulation levels of SfPelo and P7-1 in dsSfPelo-treated viruliferous insects were significantly lower than that in dsGFP-treated controls (Fig. 4c, d). In addition, at 48 hpi, P7-1 formed tubular structures in the cytoplasm of Sf9 cells treated with dsGFP. However, no tubular structures were formed until 72 hpi in the cytoplasm of Sf9 cells treated with dsSf9Pelo (Additional file 1: Figure S1). These results indicate that the knockdown of SfPelo expression by RNA interference also suppresses SRBSDV propagation in insect vectors.
Knockdown SfPelo inhibits SRBSDV propagation in S. furcifera. a Immunofluorescence assay showing the association of SRBSDV P7-1 with SfPelo in SRBSDV-infected midgut. At 6 days padp, the intestines of nonviruliferous and viruliferous S. furcifera individuals were immunostained with P7-1-rhodamine (red) and SfPelo-FITC (green), and then examined by immunofluorescence microscopy. Panels I is the enlarged images of the boxed areas, and white arrows indicate the co-localization of SfPelo and P7-1 tubules. mg, midgut. Scale bars: 5 μm. b Immunogold labeling of SfPelo in SRBSDV-infected midgut. Insect intestines were immunolabeled with SfPelo-specific IgG as the primary antibody, followed by treatment with 15-nm gold particle-conjugated IgG as the secondary antibody. Red arrows indicate gold particles. Scale bars: 200 nm. c, d The relative transcript and accumulation levels of P7-1 and SfPelo in 30 dsGFP- or dsSfPelo-treated viruliferous insects, as determined by RT-qPCR and western blot assays. Means (± SD) from three biological replicates are shown. **, P < 0.01. Insect H3 was served as reference protein. The relative intensities of bands of different proteins were determined using ImageJ
Overexpression or knockout of OsPelo inhibits the propagation of SRBSDV in rice
To determine whether OsPelo affected SRBSDV infection in rice plants, the OsPelo gene knockout or overexpressed rice plants were generated. The OsPelo overexpression (OE-OsPelo) in transgenic rice plants was obtained using the Agrobacterium tumefaciens-mediated method. Two OE-OsPelo lines displayed the same phenotype as that of wild-type (WT) rice cv. Zhonghua 11 (ZH11) (Fig. 5a). To examine whether OsPelo overexpression altered the susceptibility of rice to SRBSDV infection, WT and OE-OsPelo plants were inoculated with viruliferous S. furcifera. It was found that two OE-OsPelo lines exhibited less severe disease symptoms than WT plants (Fig. 5a). The relative transcript and protein accumulation levels of SRBSDV P7-1 in OE-OsPelo lines were significantly lower than WT controls (Fig. 5b, c). These results indicate that overexpression of OsPelo inhibits SRBSDV propagation in rice plants.
Overexpression or knockout OsPelo inhibits SRBSDV propagation in rice. a Phenotype and disease symptoms induced by SRBSDV infection in OE-OsPelo lines (OE-OsPelo-1 and OE-OsPelo-2) and wild type rice. b The relative transcript levels of P7-1 in SRBSDV-infected OsPelo overexpression transgenic lines and wild type ZH11, as determined by RT-qPCR assays. c The accumulation levels of P7-1, P10, and Pelo-Flag in SRBSDV-infected OsPelo overexpression transgenic lines and wild type ZH11, as determined by western blot assays. d Phenotype and disease symptoms induced by SRBSDV infection in KO-OsPelo lines (KO-OsPelo-1 and KO-OsPelo-2) and wild type rice. e The accumulation levels of RPS6 in OsPelo mutant transgenic lines and wild type ZH11, as determined by western blot assays. f The relative transcript levels of P7-1 in SRBSDV-infected OsPelo mutant transgenic lines and wild type ZH11, as determined by RT-qPCR. g The accumulation levels of P7-1, P10, and OsPelo in SRBSDV-infected OsPelo mutant transgenic lines and wild type ZH11, as determined by western blot assays. Means (± SD) from three biological replicates are shown. **, P < 0.01; ***, P < 0.001. Coomassie-blue stained gel for rubisco demonstrated the loading amounts of the proteins. The relative intensities of bands of different proteins were determined using ImageJ
We then utilized CRISPR/Cas9 to disrupt the function of OsPelo by modifying its corresponding coding region in the genome of rice cv. ZH11. After mutation detection in T1 generation using RT-PCR and sequencing, two homozygous mutation lines (KO-OsPelo) including an insertion mutation or a deletion mutation were successfully obtained (Additional file 1: Figure S2b, c). Two KO-OsPelo lines displayed the similar phenotype as WT rice cv. ZH11 (Fig. 5d). Furthermore, the accumulation levels of ribosomal protein RPS6 in KO-OsPelo rice plants were significantly lower than that in WT rice cv. ZH11 (Fig. 5e), consistent with the function of Pelo as the ribosome rescue factor (Bhattacharya et al. 2010). The relative transcript levels of OsPelo in two KO-OsPelo rice plants were significantly lower than that in WT rice cv. ZH11 (Additional file 1: Figure S2e), while the relative transcript levels of PR1 and PR3, two SA pathway related genes, were upregulated in KO-OsPelo transgenic rice plants (Additional file 1: Figure S3). To examine whether the knockout of OsPelo altered the susceptibility of rice to SRBSDV, WT and KO-OsPelo plants were inoculated with viruliferous S. furcifera. It was found that two KO-OsPelo lines also exhibited less severe disease symptoms than that of the WT plant (Fig. 5d). The relative transcript and protein accumulation levels of SRBSDV P7-1 in KO-OsPelo lines were significantly lower than WT control (Fig. 5f, g). It was indicated that knockout of OsPelo inhibits the infection of SRBSDV in rice plants.
OsPelo mediated antiviral activity is a conserved strategy in rice to control infection by different plant viruses
We explored whether this action of OsPelo in rice plants was conserved among different rice viruses. The phytoreovirus RDV, a dsRNA virus, and cytorhabdovirus RSMV, a negative-strand RNA virus, are transmitted by leafhoppers in a persistent-propagative manner. We inoculated WT, OE-OsPelo, and KO-OsPelo rice plants with RDV-infected N. cincticeps or RSMV-infected R. dorsalis. Western blot assays confirmed that the protein accumulation levels of RDV protein Pns10 in OE-OsPelo and KO-OsPelo lines were significantly lower than that in WT ZH11 (Fig. 6a, b). Similarly, the protein accumulation levels of RSMV protein N in OE-OsPelo and KO-OsPelo lines were also significantly lower than WT ZH11 (Fig. 6c, d). Thus, overexpression or knockout of OsPelo inhibits the propagation of different plant viruses in rice.
Overexpression or knockout OsPelo inhibits SRBSDV propagation in rice. a, b The accumulation levels of RDV Pns10 in RDV-infected OsPelo overexpression and OsPelo mutant transgenic lines and wild type ZH11, as determined by western blot assays. c, d The accumulation levels of RSMV N in RSMV-infected OsPelo overexpression and OsPelo mutant transgenic lines and wild type ZH11, as determined by western blot assays. Coomassie-blue stained gel for rubisco demonstrated the loading amounts of the proteins. The relative intensities of bands of different proteins were determined using ImageJ
Discussion
Plants and insects have evolved mRNA quality control systems to ensure the proper synthesis of their own proteins (D’ Orazio and Green 2021). Recently, a few examples have described the interplay between NGD and plant viruses (Ren et al. 2022; Ge et al. 2023). It has been well established that Hbs1-Pelo complex in NGD system functions as a regulator to dissociate stalled ribosomes of aberrant mRNAs (Saito et al. 2013; Kong et al. 2021). When confronted with viral infection, the cell might recognize virus, and potentially initiates Pelo-Hbs1 complex to deal with abnormal ribosomes and viral mRNAs to inhibit viral infection. Our results show that overexpression of OsPelo in transgenic rice increases the resistance to infection of the fijivirus SRBSDV. Furthermore, microinjection of purified SfPelo in S. furcifera also inhibits effective SRBSDV propagation. SRBSDV employs virus-induced P7-1 tubular structure as a vehicle for viral spread throughout the body of S. furcifera (Jia et al. 2014). The co-expression of SfPelo with P7-1 could inhibit the assembly of P7-1 tubular structures in Sf9 cells. More importantly, the overexpression of OsPelo in transgenic rice displays broad-spectrum resistance to infection of the phytoreovirus RDV and cytorhabdovirus RSMV. Because both OsPelo and SfPelo could directly interact with P7-1 and their GTPase partner Hbs1, we thus deduce that Pelo-Hbs1 complex serves as the defender against rice viruses in rice plants and insect vectors.
Pelo is the key factor recognizing ribosomes that have stalled during elongation, and functioning in mRNA quality control process (Ikeuchi et al. 2016; Szádeczky-Kardoss et al. 2018). Increasing studies have found that knockout or mutation of Pelo can improve the antiviral properties of host. Drosophila knockout mutants of Pelo show resistance against several viruses (Wu et al. 2014). Tomato knockout mutants of Pelo also have broad-spectrum resistance to geminiviruses (Lapidot et al. 2015). In this study, the effective SRBSDV propagation is also inhibited in the OsPelo knockout transgenic rice plants. Ribosomal proteins play a significant role in mRNA quality control process. In general, ribosomal proteins play a crucial role in the lifecycle of certain viruses, and viruses could exploit the cellular translation system to promote their propagation (Fukushi et al. 2001; Simms et al. 2017; Li et al. 2018). It has been reported that, the deletion of yeast strains of dom34 (homology of Pelo in yeast) leads to the increased non-functional 80S monosomes and decreased polysomes (Bhattacharya et al. 2010; van den Elzen et al. 2014). In this study, the accumulation levels of ribosomal protein S6 (RPS6), a part of the small ribosomal subunit, is significantly reduced in OsPelo-knockout transgenic rice plants. Furthermore, the knockout of OsPelo in transgenic rice exhibits broad-spectrum resistance to the fijivirus SRBSDV, phytoreovirus RDV, and cytorhabdovirus RSMV infection. The knockdown of SfPelo expression in S. furcifera also shows resistance to SRBSDV infection. These phenomena suggest that the absence or the down-regulation of Pelo expression affects the expression of ribosomal proteins and thus adversely affects the propagation of viruses. It is presumed that the excessive inhibition of Pelo expression in rice plants or insect vectors not only weakens the mRNA surveillance system but also reduces the availability of free ribosomes within the cell, thus limiting the synthesis of viral proteins and viral replication.
To ensure viral persistent infection, virus employs some strategies to counteract mRNA quality control mechanisms (Ge et al. 2023). In our study, we discover that the accumulation levels of OsPelo and SfPelo are slightly decreased in SRBSDV infected rice and S. furcifera, thus weakening the mRNA surveillance system of the two viral hosts. Such moderate reduction of Pelo accumulation during vial propagation in plant hosts and insect vectors would avoid Pelo-Hbs1-mediated excessive inhibition of P7-1 tubular structure formation, ensuring effective virus propagation. It would be valuable to investigate whether P7-1 or other virus-encoded proteins modulate the reduced expression of Pelo during viral infection in insect vectors or rice plants.
It has been reported that the natural mutants of the pepy-1 gene in pepper and ty-5 gene in tomato encoding Pelo protein confer resistance to begomovirus isolates by restricting virus replication to a level virus tolerance than an immune response (Koeda et al. 2021). OsPelo mutant rice with a single base substitution from T to A at position 556 shows a spotted-leaf phenotype and highly upregulated the pathogenesis-related protein marker genes to enhance the resistance to rice bacterial blight (Ding et al. 2018; Zhang et al. 2018). In our study, the OsPelo knockout transgenic rice shows mild symptoms and a low level of SRBSDV accumulation in rice leaves, which corresponds to the phenomenon in other Pelo-resistant lines (Koeda et al. 2021). Meanwhile, the expression levels of two SA pathway related genes, PR1 and PR3, were significantly upregulated in the OsPelo knockout transgenic rice, which indicating the SA pathway were activated and might play a role in enhancing pathogen resistance (Ding et al. 2018; Zhang et al. 2018). Especially, OsPelo-knockout transgenic plants limit the propagation of different rice viruses, such as RDV and RSMV in rice plants. Generally, the selection of a resistant variety is an effective method for controlling plant virus diseases. Therefore, screening the natural mutants of OsPelo gene in different rice germplasm resources and identifying its resistance to rice viruses are feasible to produce resistant materials.
Methods
Insects, viruses, and antibodies
Nonviruliferous S. furcifera was collected from rice fields in Zhangzhou city, Fujian Province and reared on TN-1 rice seedlings at 25 ± 1 °C with 75 ± 5% relative humidity and 16 h light/8 h dark. The SRBSDV infected plants were collected from Zhangzhou city and propagated by viruliferous S. furcifera. The Sf9 cell line was cultured in growth medium Sf900 III (Gbico). Rabbit polyclonal antibodies against SfPelo of S. furcifera, OsPelo of rice, P7-1, and P10 of SRBSDV were prepared as previously described (Jia et al. 2014). Mouse monoclonal antibody against GST was purchased from Transgene Biotech (HT601). Mouse polyclonal antibody against H3 was purchased from Proteintech (17168-1-AP). Mouse monoclonal antibody against 6×His tag was purchased from Sangon Biotech (D191001).
Y2H assay
To investigate the interactions among P7-1, Pelo, and Hbs1, the ORFs of SRBSDV P7-1, OsPelo, OsHbs1, SfPelo, and SfHbs1 were individually amplified and cloned into the bait plasmid pGBKT7 or the prey plasmid pGADT7. The primers for Y2H assay were listed in Additional file 2: Table S1. The bait and prey plasmids were used to co-transform the yeast strain AH109 according to the manufacturer’s instructions (Clontech), and the transformants were grown on SD/-Leu-Trp (DDO) and SD/-Leu-Trp-His-Ade (QDO) plates. Yeast cells were photographed at 30 °C to record growth. The pGBKT7-53/pGADT7-T served as positive control and pGBKT7-Lam/pGADT7-T served as negative control.
RT-qPCR assay
Total RNAs of rice leaves, S. furcifera or Sf9 cells were extracted using TRIzol reagent (Invitrogen). The RT-qPCR assays were performed in the QuantStudio™5 Real-Time PCR System (Applied Biosystem) using the SYBR Green PCR Master Mix Kit (GenStar). Relative expression levels of P7-1, OsPelo, and SfPelo were analyzed using the 2−ΔΔCT method (Livak and Schmittgen 2001). The housekeeping gene actin of Oryza sativa, and EF1 of S. furcifera were served as the internal reference for the normalization of gene expression levels. The primers used for RT-qPCR assays were shown in Additional file 2: Table S1. All RT-qPCRs were conducted in triplicate for each sample, and three biological replicates were maintained.
Western blot assay
To analyze the accumulation levels of viral proteins, OsPelo or SfPelo in rice plants, S. furcifera or Sf9 cells, total proteins were extracted and separated on 12% SDS-PAGE gels before transfer to the pre-activated PVDF membrane. Antibodies against SRBSDV P7-1, P10, OsPelo, and SfPelo were used as the primary antibodies and goat anti-rabbit IgG-peroxidase (Sigma-Aldrich) was used as the secondary antibody. The accumulation level of H3 served as the reference protein of S. furcifera using H3-specific antibody (Proteintech). The accumulation level of GAPDH served as the reference protein of Sf9 cells using GAPDH-specific antibody (Sangon Biotech). Coomassie-blue stained gel for rubisco demonstrated the loading amounts of the proteins of rice plants. The proteins were visualized using the Luminata Classico Western HRP Substrate (Millipore) and were imaged using the Molecular Imager ChemiDoc XRS + System (Bio-Rad). ImageJ software (https://imagej.nih.gov/ij/) was used for measuring band intensities of proteins.
GST pull-down
The ORF of P7-1 was inserted into the pGex-4T-3 vector to generate plasmids expressing GST fusion protein. The ORFs of OsPelo and SfPelo were cloned into the pET-28a to construct the plasmid expressing His-fusion protein. The primers used for GST pull-down assay listed in Additional file 2: Table S1. The recombinant protein of GST-P7-1 and GST were expressed in E. coli stain BL21. Lysates were then incubated with glutathione-Sepharose beads (Amersham) and subsequently with the recombinant proteins His-OsPelo or His-SfPelo which was also expressed in E. coli stain BL21, washing with 1×PBS in each step. Finally, the immunoprecipitated proteins were analyzed by western blot assay using GST-tag and His-tag antibodies, respectively.
Effect of synthesized dsRNA or protein on viral accumulation
The dsRNA targeting Pelo of S. furcifera (dsSfPelo) and Spodoptera frugiperda 9 cells (dsSf9Pelo) was synthesized in vitro using the T7 RiboMAX Express RNAi System (Promega Biotech, P1700), according to the manufacturer’s protocol. To test the effect of dsSfPelo on SRBSDV accumulation, 200 third-instar nymphs of S. furcifera were fed with SRBSDV-infected rice for 2 days. Then, 0.5 µg/µL dsRNAs were microinjected into the S. furcifera, followed by transferring to healthy rice seedlings. The dsGFP treatment served as control. At 5 days padp, the relative transcript levels of SfPelo and SRBSDV P7-1 was examined by RT-qPCR and western blot assay as described above. The primers used for the RNAi assay are listed in Additional file 2: Table S1. All the experiments were repeated three times. Insect H3 was served as the reference protein. ImageJ software (https://imagej.nih.gov/ij/) was used for signal quantitation.
To confirm the role of SfPelo in the suppression of the propagation of SRBSDV in vivo, the SfPelo and GFP proteins were expressed and purified following the methods described above (Jia et al. 2023). Approximately 200 third instar nymphs of S. furcifera were allowed to feed on infected rice plants for 2 days. Viruliferous S. furcifera individuals were then microinjected with purified SfPelo or GFP at a concentration of 300 µg/mL. At 4 days post-microinjection, the insects were tested for the accumulation of P7-1 protein using western blot assays, as described above.
Baculovirus expression assay
P7-1, Sfpelo, and GFP were respectively expressed in a baculovirus according to the manufacturer’s instructions (Thermo Fisher Scientific). We amplified and cloned DNA sequences encoding His-tagged SfPelo (SfPelo-His), P7-1, and GFP into pFast-bac1 vector to construct recombinant baculoviruses. The primers used for bacmid expression assay are listed in Additional file 2: Table S1. The procedures of bacmids transfection were performed according to the manufacturer’s instructions (Thermo Fisher Scientific). To observe the effect of SfPelo on P7-1, recombinant bacmids expressing SfPelo-His was co-infected with GFP or P7-1 into Sf9 cells. At 24, 48, and 72 hpi, the cells were fixed, permeabilized, immunolabeled with antibody against P7-1 conjugated to rhodamine (P7-1-rodamine) or His conjugated to FITC (His-FITC), and then processed for immunofluorescence microscopy. Furthermore, the Sf9 cells infected with bacmids were collected for western blot and RT-qPCR assays, as described above. To test the effect of dsSf9Pelo on tubular formation of P7-1, the Sf9 cells treated with dsSf9Pelo or dsGFP for 6 h were infected with recombinant bacmids containing P7-1. At 48, 72 hpi, then the cells were fixed, permeabilized, immunolabeled with antibody against P7-1 conjugated to rhodamine (P7-1-rodamine), before being processed for immunofluorescence microscopy.
OsPelo overexpressed and knockout transgenic rice lines
The full length sequence encoding OsPelo fused with Flag-tag was cloned, followed by the construction of the expression vector through homologous recombination to generate transgenic rice plants overexpressing OsPelo at Biorun Bio-Company (Wuhan, China). The recombinant plasmid was electroporated into Agrobacterium tumefaciens strain GV3101 and used to transform into rice cv. Zhonghua 11 (ZH11). The T1 generation overexpressed transgenic lines that stably maintained the transgenes were determined by western blot assays and selected for phenotype analyses. The T0 generation OsPelo knock-out CRISPR/Cas9-editted rice cv. ZH11 targeting the sequence GGATCTGTCTTGCGTGTACGTGG were obtained using the Agrobacterium-mediated method at Biorun Bio-Company (Wuhan, China). In the T0 generation population, exogenous transgenic components were detected by sequencing and PCR assays using specific primers targeting OsPelo and the hygromycin gene to obtain the mutation lines. In the T1 generation population, exogenous transgenic components were detected by sequencing and RT-PCR assays using specific primers targeting OsPelo-encoding sequence to obtain steady homozygous T2 lines for further experiments. The primers used for detection of the knockout transgenic rice plants are listed in Additional file 2: Table S1.
Virus inoculation assays
To inoculate virus into transgenic rice, 100 third-instar insects were allowed to feed continuously on SRBSDV-, RDV-, or RSMV-infected rice plants for 3 days, and then transferred on healthy rice seedlings for 10 days. Subsequently, the third leaf stage transgenic and wild type rice seedlings were fed with viruliferous insects for 2 days. Then the insects were removed and rice seedlings were grown in the field for symptom development. At 7 dpi, the viruliferous rice plants were detected by PCR and the accumulation levels of SRBSDV P7-1, RDV Pns10, or RSMV N were detected by western blot assays, as described above. The disease symptoms were observed in about three months.
Immunofluorescence and immunoelectron microscopy
To visualize the association of SfPelo with P7-1 of SRBSDV in the midgut, the intestines from 30 nonviruliferous and viruliferous S. furcifera, were dissected. The samples were fixed in 4% (v/v) paraformaldehyde in phosphate belanced solution (PBS) for 2 h, and then permeabilized in 0.2% (v/v) Triton-X for 1 h. The samples were then immunolabeled with antibody against P7-1 directly conjugated to rhodamine (P7-1-rhodamine), and antibody against SfPelo directly conjugated to FITC (SfPelo-FITC) (0.5 µg/µL). Immunostained tissues were analyzed using a Leica TCS SPE inverted confocal microscope. Meanwhile, the intestines from nonviruliferous and viruliferous S. furcifera insects were fixed, dehydrated and embedded as described previously (Mao et al. 2013). Polymerization was allowed to proceed for 72 h at -20°C. The samples were then sectioned, and the ultrathin sections were immunolabeled with SfPelo antibody (0.5 µg/µL) as the primary antibody, goat anti-rabbit IgG conjugated with 15-nm diameter gold particles (0.5 µg/µL, Sigma-Aldrich) as the secondary antibody. Finally, the ultrathin sections were analyzed using a transmission electron microscope (H-7650, Hitachi).
Sequence homology analysis
The amino acid sequence encoded by SfPelo or OsPelo was predicted by online software of Smart (http://smart.embl-heidelberg.de/). The aino acid sequences of SfPelo and OsPelo were aligned using the software DNAMAN 9. Phylogenetic tree of Pelo proteins from different species was constructed by Neighbor-Joining method using the software MEGA 6.0.
Availability of data and materials
The data and materials that support the findings of this study are available from the corresponding author upon request.
Abbreviations
- aa:
-
Amino acid
- BNIP3:
-
BCL2 interacting protein 3
- DDO:
-
SD/-Leu-Trp
- dsRNA:
-
Double-stranded RNA
- ER:
-
Endoplasmic reticulum
- GFP:
-
Green fluorescent protein
- GST:
-
Glutathione-S-transferase
- Hbs1:
-
Hsp70 subfamily B suppressor 1
- hpi:
-
Hours post-infection
- KO:
-
Knockout
- LAMP1:
-
Lysosomal-associated membrane protein 1
- mRNA:
-
Messenger RNA
- NGD:
-
No-go mRNA decay
- NMD:
-
Nonsense-mediated decay
- NSD:
-
No-stop decay
- ORF:
-
Open reading frame
- PBS:
-
Phosphate belanced solution
- Pelo:
-
Pelota
- PR1:
-
Pathogenesis-related protein 1
- PR3:
-
Pathogenesis-related protein 3
- QDO:
-
SD/-Leu-Trp-His-Ade
- RDV:
-
Rice dwarf virus
- RGDV:
-
Rice gall dwarf virus
- RPS6:
-
Ribosomal protein S6
- RQC:
-
RNA quality control
- RSMV:
-
Rice stripe mosaic virus
- RT-qPCR:
-
Real-Time quantitative polymerase chain reaction
- SA:
-
Salicylic acid
- Sf9:
-
Spodoptera frugiperda 9
- SRBSDV:
-
Southern rice black-streaked dwarf virus
- WT:
-
Wild-type
- Y2H:
-
Yeast two-hybrid
- ZH11:
-
Zhonghua 11
References
Aly HH, Suzuki J, Watashi K, Chayama K, Hoshino S, Hijikata M, et al. RNA exosome complex regulates stability of the Hepatitis B virus X-mRNA transcript in a non-stop-mediated (NSD) RNA quality control mechanism. J Biol Chem. 2016;291(31):15958–74. https://doi.org/10.1074/jbc.M116.724641.
Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol. 2010;30(23):5562–71. https://doi.org/10.1128/MCB.00618-10.
Bicknell AA, Ricci EP. When mRNA translation meets decay. Biochem Soc Trans. 2017;45(2):339–51. https://doi.org/10.1042/BST20160243.
Chen Q, Chen H, Mao Q, Liu Q, Shimizu T, Uehara-Ichiki T, et al. Tubular structure induced by a plant virus facilitates viral spread in its vector insect. PLoS Pathog. 2012;18(11):e1003032. https://doi.org/10.1371/journal.ppat.1003032.
Ding W, Wu J, Ye J, Zheng W, Wang S, Zhu X, et al. A pelota-like gene regulates root development and defence responses in rice. Ann Bot. 2018;122(3):359–71. https://doi.org/10.1093/aob/mcy075.
D’Orazio KN, Green R. Ribosome states signal RNA quality control. Mol Cell. 2021;81(7):1372–83. https://doi.org/10.1016/j.molcel.2021.02.022.
Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276(24):20824–6. https://doi.org/10.1074/jbc.C100206200.
Ge L, Cao B, Qiao R, Cui H, Li S, Shan H, et al. SUMOylation-modified Pelota-Hbs1 RNA surveillance complex restricts the infection of potyvirids in plants. Mol Plant. 2023;16(3):632–42. https://doi.org/10.1016/j.molp.2022.12.024.
Huang X, Wang J, Chen S, Liu S, Li Z, Wang Z, et al. Rhabdovirus encoded glycoprotein induces and harnesses host antiviral autophagy for maintaining its compatible infection. Autophagy. 2023;1:1–20. https://doi.org/10.1080/15548627.2023.2252273.
Ikeuchi K, Yazaki E, Kudo K, Inada T. Conserved functions of human Pelota in mRNA quality control of nonstop mRNA. FEBS Lett. 2016;590(18):3254–63. https://doi.org/10.1002/1873-3468.12366.
Jamar NH, Kritsiligkou P, Grant CM. Loss of mRNA surveillance pathways results in widespread protein aggregation. Sci Rep. 2018;8(1):3894. https://doi.org/10.1038/s41598-018-22183-2.
Jia D, Mao Q, Chen H, Wang A, Liu Y, Wang H, et al. Virus-induced tubule: a vehicle for rapid spread of virions through basal lamina from midgut epithelium in the insect vector. J Virol. 2014;88(18):10488–500. https://doi.org/10.1128/jvi.01261-14.
Jia D, Mao Q, Chen Y, Liu Y, Chen Q, Wu W, et al. Insect symbiotic bacteria harbours viral pathogens for transovarial transmission. Nat Microbiol. 2017;2:17025. https://doi.org/10.1038/nmicrobiol.2017.25.
Jia D, Luo G, Shi W, Liu Y, Liu H, Zhang X, et al. Rice gall dwarf virus promotes the propagation and transmission of rice stripe mosaic virus by co-infected insect vectors. Front Microbiol. 2022;13:834712. https://doi.org/10.3389/fmicb.2022.834712.
Jia D, Liang Q, Chen H, Liu H, Li G, Zhang X, et al. Autophagy mediates a direct synergistic interaction during co-transmission of two distinct arboviruses by insect vectors. Sci China Life Sci. 2023;66:1665–81. https://doi.org/10.1007/s11427-022-2228-y.
Koeda S, Onouchi M, Mori N, Pohan NS, Nagano AJ, Kesumawati E. A recessive gene pepy-1 encoding Pelota confers resistance to begomovirus isolates of PepYLCIV and PepYLCAV in Capsicum annuum. Theor Appl Genet. 2021;134(9):2947–64. https://doi.org/10.1007/s00122-021-03870-7.
Kong W, Tan S, Zhao Q, Lin D, Xu Z, Friml J, et al. mRNA surveillance complex PELOTA-HBS1 regulates phosphoinositide-dependent protein kinase1 and plant growth. Plant Physiol. 2021;186(4):2003–20. https://doi.org/10.1093/plphys/kiab199.
Lapidot M, Karniel U, Gelbart D, Fogel D, Evenor D, Kutsher Y, et al. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genet. 2015;11(10):e1005538. https://doi.org/10.1371/journal.pgen.1005538.
Lee HH, Kim YS, Kim KH, Heo I, Kim SK, Kim O, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell. 2007;27(6):938–50. https://doi.org/10.1016/j.molcel.2007.07.019.
Li F, Wang A. RNA-targeted antiviral immunity: more than just RNA silencing. Trends Microbiol. 2019;27:792–805. https://doi.org/10.1016/j.tim.2019.05.007.
Li S, Li X, Zhou Y. Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res. 2018;247:15–20. https://doi.org/10.1016/j.virusres.2018.01.011.
Li Z, Zhang T, Huang X, Zhou G. Impact of two reoviruses and their coinfection on the rice RNAi system and vsiRNA production. Viruses. 2018b;10(11):594. https://www.mdpi.com/1999-4915/10/11/594.
Liang Q, Wan J, Liu H, Chen M, Xue T, Jia D, et al. A plant reovirus hijacks the DNAJB12-Hsc70 chaperone complex to promote viral spread in its planthopper vector. Mol Plant Pathol. 2022;23(6):805–18. https://doi.org/10.1111/mpp.13152.
Liang Q, Wan J, Liu H, Jia D, Chen Q, Wang A, et al. A plant nonenveloped double-stranded RNA virus activates and co-opts BNIP3-mediated mitophagy to promote persistent infection in its insect vector. Autophagy. 2023;19(2):616–31. https://doi.org/10.1080/15548627.2022.2091904.
Liu Y, Jia D, Chen H, Chen Q, Xie L, Wu Z, et al. The P7-1 protein of southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch Virol. 2011;156(10):1729–36. https://doi.org/10.1007/s00705-011-1041-9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods. 2001;25:402–8. https://doi.org/10.1006/meth.2001.1262.
Mao Q, Zheng S, Han Q, Chen H, Ma Y, Jia D, et al. New model for the genesis and maturation of viroplasms induced by fijiviruses in insect vector cells. J Virol. 2013;87(12):6819–28. https://doi.org/10.1128/JVI.00409-13.
Nogimori T, Nishiura K, Kawashima S, Nagai T, Oishi Y, Hosoda N, et al. Dom34 mediates targeting of exogenous RNA in the antiviral OAS/RNase L pathway. Nucleic Acids Res. 2019;47(1):432–49. https://doi.org/10.1093/nar/gky1087.
Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, et al. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20(13):3025–32. https://doi.org/10.1091/mbc.e09-01-0028.
Ren Y, Tao X, Li D, Yang X, Zhou X. ty-5 Confers broad-spectrum resistance to geminiviruses. Viruses. 2022;14(8):1804. https://doi.org/10.3390/v14081804.
Saito S, Hosoda N, Hoshino S. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J Biol Chem. 2013;288(24):17832–43. https://doi.org/10.1074/jbc.M112.448977.
Simms CL, Thomas EN, Zaher HS. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev RNA. 2017;8(1). https://doi.org/10.1002/wrna.1366.
Szádeczky-Kardoss I, Gál L, Auber A, Taller J, Silhavy D. The No-go decay system degrades plant mRNAs that contain a long A-stretch in the coding region. Plant Sci. 2018;275:19–27. https://doi.org/10.1016/j.plantsci.2018.07.008.
Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, et al. Dom34: hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3’ end of aberrant mRNA. Mol Cell. 2012;46:518–29. https://doi.org/10.1016/j.molcel.2012.03.013.
van den Elzen AM, Schuller A, Green R, Séraphin B. Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. EMBO J. 2014;33(3):265–76. https://doi.org/10.1002/embj.201386123.
van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends Biochem Sci. 2011;36(11):585–92. https://doi.org/10.1016/j.tibs.2011.07.005.
Wang Y, Jiang J, Zhao L, Zhou R, Yu W, Zhao T. Application of whole genome resequencing in mapping of a tomato yellow leaf curl virus resistance gene. Sci Rep. 2018;8(1):9592. https://doi.org/10.1038/s41598-018-27925-w.
Wu X, He W, Tian S, Meng D, Li Y, Chen W, et al. Pelo is required for high efficiency viral replication. PLoS Pathog. 2014;10(4):e1004034. https://doi.org/10.1371/journal.ppat.1004034.
Yang X, Huang J, Liu C, Chen B, Zhang T, Zhou G. Rice stripe mosaic virus, a novel cytorhabdovirus infecting rice via leafhopper transmission. Front Microbiol. 2017;7:2140. https://doi.org/10.3389/fmicb.2016.02140.
Yang X, Chen B, Zhang T, Li Z, Xu C, Zhou G. Geographic distribution and genetic diversity of rice stripe mosaic virus in Southern China. Front Microbiol. 2018;9:3068. https://doi.org/10.3389/fmicb.2018.03068.
Yin X, Xu F, Zheng F, Li X, Liu B, Zhang C. Molecular characterization of segments S7 to S10 of a southern rice black-streaked dwarf virus isolate from maize in northern China. Virol Sin. 2011;26(1):47–53. https://doi.org/10.1007/s12250-011-3170-9.
Yu X, Jia D, Wang Z, Li G, Chen M, Liang Q, et al. A plant reovirus hijacks endoplasmic reticulum-associated degradation machinery to promote efficient viral transmission by its planthopper vector under high temperature conditions. PLoS Pathog. 2021;17(3):e1009347. https://doi.org/10.1371/journal.ppat.1009347.
Zhang X, Feng B, Wang H, Xu X, Shi Y, He Y, et al. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J Integr Plant Biol. 2018;60(2):160–72. https://doi.org/10.1111/jipb.12613.
Zhang L, Liu W, Wu N, Wang H, Zhang Z, Liu Y, et al. Southern rice black-streaked dwarf virus induces incomplete autophagy for persistence in gut epithelial cells of its vector insect. PLoS Pathog. 2023;19(1):e1011134. https://doi.org/10.1371/journal.ppat.1011134.
Zhao Y, Ye X, Shehata M, Dunker W, Xie Z, Karijolich J. The RNA quality control pathway nonsense-mediated mRNA decay targets cellular and viral RNAs to restrict KSHV. Nat Commun. 2020;11:3345. https://doi.org/10.1038/s41467-020-17151-2.
Zhou G, Xu D, Xu D, Zhang M. Southern rice black-streaked dwarf virus: a new proposed fijivirus species in the family Reoviridae. Chin Sci Bull. 2008;53:3677–85. https://doi.org/10.1007/s11434-008-0467-2.
Zhou G, Xu D, Xu D, Zhang M. Southern rice black-streaked dwarf virus: a white-backed planthopper-transmitted fijivirus threatening rice production in Asia. Front Microbiol. 2013;4:270. https://doi.org/10.3389/fmicb.2013.00270.
Zhou S, Zhao Y, Liang Z, Wu R, Chen B, Zhang T, Yang X, Zhou G. Resistance evaluation of dominant varieties against southern rice black-streaked dwarf virus in southern China. Viruses. 2021;13(8):1501. https://www.mdpi.com/1999-4915/13/8/1501.
Acknowledgements
We thank members of the Wei lab for stimulating discussions and technical assistance.
Funding
This project was supported by funds from the National Natural Science Foundation of China (31920103014 and U23A20197).
Author information
Authors and Affiliations
Contributions
TW, XS, and DJ designed all experiments. XS and DJ performed the experiments for immunofluorescence staining and electron microscopy. XS and HG performed the protein interaction experiments. DJ and TW analyzed the data. TW, DJ, and XS organized the project and wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1: Figure S1.
Knockdown Sf9-Pelo inhibits the tubular formation of P7-1 in Sf9 cells. Figure S2. Detection of the OsPelo mutant transgenic rice plants. Figure S3. Detection of the SA signal pathway genes in OsPelo mutant transgenic rice plants.
Additional file 2: Table S1.
The primers used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, X., Guan, H., Wei, T. et al. The mRNA surveillance factor Pelo restricts rice virus propagation in insect vectors and host plants. Phytopathol Res 6, 34 (2024). https://doi.org/10.1186/s42483-024-00251-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-024-00251-y