Abstract
Wheat dwarf virus (WDV), a mastrevirus transmitted by the leafhopper Psammotettix alienus, causes a severe disease in cereal crops. Typical symptoms of wheat plants infected by WDV are yellowing and severe dwarfing. In this present study, RNA-Seq was used to perform gene expression analysis in wheat plants in response to WDV infection. Comparative transcriptome analysis indicated that a total of 1042 differentially expressed genes (DEGs) were identified in the comparison between mock and WDV-inoculated wheat plants. Genomes ontology (GO) annotation revealed a number of DEGs associated with different biological processes, such as phytohormone metabolism, photosynthesis, DNA metabolic process, response to biotic stimulus and defense response. Among these, DEGs involved in phytohormone and photosynthesis metabolism and response pathways were further enriched and analyzed, which indicated that hormone biosynthesis, signaling and chloroplast photosynthesis-related genes might play an important role in symptom development after WDV infection. These results illustrate the dynamic nature of the wheat-WDV interaction at the transcriptome level and confirm that symptom development is a complex process, providing a solid foundation to elucidate the pathogenesis of WDV.
Similar content being viewed by others
Background
Wheat dwarf disease (WDD) causes serious yield losses in cereal crops including wheat and barley worldwide, particularly in Europe and Asia (Vacke 1961; Lindsten and Lindste 1999; Najar et al. 2000; Xie et al. 2007). Yield losses may reach up to 100% and have been reported to occur due to severe infections in winter cereals (wheat/barley) in some regions of Germany, Hungary, Czech Republic and China (Bisztray et al. 1989; Commandeur and Huth 1999; Kundu et al. 2009; Liu et al. 2012). Symptoms of WDD typically include extremely dwarfed plant height, yellowing or streaking on leaves, and even failure of heading (Lindblad and Sigvald 2004). WDD is caused by the wheat dwarf virus (WDV), which is transmitted in a persistent manner by the leafhopper Psammotettix alienus (Vacke 1961; Wang et al. 2019a). WDV belongs to the genus Mastrevirus in the family Geminiviridae and its genome comprises of a monopartite, single-stranded (ss), circular DNA (Gutierrez 1999). The genome of WDV encodes four proteins: the capsid protein (CP) and the movement protein (MP) on the viral-sense strand and two replication associated proteins (Rep and RepA) on the complementary-sense strand (Gutierrez 1999). The Rep protein, a RNA silencing suppressor, also has been demonstrated to be involved in pathogenicity (Liu et al. 2014). However, the molecular mechanisms (pathways) occurring during WDV-wheat interactions that result in symptom development are still unknown.
Phytohormones are involved in all plant growth and developmental processes. They also play an important role in plant defense response to invading microbial pathogens (Berens et al. 2017). Traditionally, plant hormones include auxin (AUX), gibberellins (GAs), cytokinins (CKs), abscisic acid (ABA), ethylene (ET), jasmonic acid (JA), salicylic acid (SA), and brassinosteroids (BRs) (Weyers and Paterson 2001). Among these, eight signaling molecules (AUX, ABA, CKs, ET, GAs, SA, JA and BRs) are reported to play key roles in mediating disease resistance (Koornneef and Pieterse 2008; De Vleesschauwer et al. 2014). Plant hormone-regulated systems are also involved in the defense against viral infections (Sanchez et al. 2010; Alazem and Lin 2015). The complex interactions between plants and viruses often lead to the appearance of symptoms such as stunting of growth, yellowing and mottling on leaves (Fraser and Whenham 1982). To some extent, symptoms caused by a viral infection may be simplistically attributed to the change in quantity of a certain plant hormone, and can be mimicked by application or removal of this plant hormone (Jameson and Clarke 2002). For example, rice plants infected with rice dwarf virus (RDV) exhibit a dwarf phenotype, which correlates to reduced levels of GA1. However, these defects were rescued by exogenous application of GA3 (Zhu et al. 2005). Furthermore, the BR pathway is associated with susceptibility whereas the JA pathway is involved in resistance to rice black-streaked dwarf virus (RBSDV) (He et al. 2017).
Photosynthesis is another biological activity that seems to be repressed during plant virus infection (Adolph and Haselkorn 1972). Some viral infections have an impact on the host’s photosynthesis, which can be associated with viral symptoms of leaf chlorosis and mosaic (Guo et al. 2005; Funayama-Noguchi and Terashima 2006; Song et al. 2009). For example, molecular studies in Nicotiana benthamiana plants infected with pepper mild mottle virus (PMMoV) revealed that several proteins involved in both the photosynthetic electron-transport chain and the Benson-Calvin cycle are down-regulated during viral infection (Pineda et al. 2010). Similarly, proteomic analyses of maize (Zea mays) seedlings in response to sugarcane mosaic virus (SCMV) infection revealed that most of the photosynthesis-related proteins were down-regulated with the exception of the RuBisCO large subunit and the ferredoxin-NADP reductase and its isoforms (Wu et al. 2013). Moreover, it is suggested that chloroplast photosynthesis-related genes/proteins (CPRGs/CPRPs) play important roles during the complex interaction between plant and virus (Zhao et al. 2016). Thus, the disruption of chloroplast structure and function may be a key element in the production of chlorosis symptoms (Balachandran et al. 1997; Manfre et al. 2011). Indeed, increasing evidence suggests that the chloroplasts are a common target of plant viruses and are involved in viral symptom development (Díaz-Vivancos et al. 2008; Lu et al. 2012).
Recently, next-generation deep sequencing-based transcriptomics has been used for studying comprehensive changes in host gene expression during plant-virus interactions (Jia et al. 2012; Allie et al. 2014; Zhou et al. 2016). Furthermore, transcriptomics has also been applied to identify the specific metabolic changes that are associated with symptom expression, which correspond to pathological and physiological changes in the host plants (Hanssen et al. 2011; Rizza et al. 2012; Mochizuki et al. 2014). In this study, we analyzed the transcriptome of wheat plants infected by WDV through mRNA expression by RNA-Seq. The results provide further insights into the molecular mechanisms of WDV pathogenesis during plant-virus interactions.
Results
Sequencing and de novo assembly of transcriptome
To obtain a global view of the transcriptome changes of the wheat plants in response to WDV infection, the expression profiles of WDV-infected wheat (W20–1, W20–2 and W20–3) were compared to mock-inoculated control plants (CK-1, CK-2 and CK-3) by high-throughput sequencing. RNA-seq yielded 46,590,920 to 53,178,180 and 54,167,714 to 60,571,914 raw reads for the WDV-infected plant group (WH20–1, W20–2 and W20–3) and the control group (CK-1, CK-2 and CK-3), respectively. After the low-quality reads and adapter sequences were removed, the clean reads were ranged from 43,435,900 to 50,447,680 for W20 group, and 51,188,492 to 56,991,586 for CK group (Table 1).
Identification of differentially expressed genes (DEGs) in wheat plants responding to WDV
Gene mapping rate was between 92.92% and 95.28% (Table 1). Differentially expressed genes responding to WDV infection were identified by comparing the W20 group to the CK group using DESeq2. Padj < 0.05 was used as the threshold to screen the DEGs. Finally, 1042 genes of the W20 group were differentially expressed. Among these genes, 465 (44.6%) were up-regulated, whereas 577 (55.4%) were down-regulated.
Functional annotation and classification
GO assignment was performed to classify the gene function of unigenes. Both up-regulated and down-regulated DEGs can be categorized into three main categories: biological process, cellular component and molecular function (Fig. 1). For down-regulated DEGs, in each of the three main categories of the GO classification, ‘cell redox homeostasis’, ‘oxidoreductase activity, acting on a sulfur group of donors’ and ‘chloroplast part’ were the significantly enriched terms; while for up-regulated DEGs, the most significantly enriched terms were ‘nucleic acid metabolic process’, ‘nucleic acid binding transcription factor activity’ and ‘mismatch repair complex’. We also noticed a high-percentage of genes from terms involved in ‘single-organism process’, ‘plastid’ and ‘catalytic activity’.
Classification of gene ontology (GO) analysis of selected DEGs after inoculation with WDV. a Top 30 GO terms enriched for down-regulated DEGs. b Top 30 GO terms enriched for up-regulated DEGs. The y-axis represents the enriched GO term, and the x-axis represents the number of DEGs in the term. Different colors are used to distinguish biological processes, cellular components, and molecular functions. GO term with asterisk means significant enrichment
To further understand the biological processes in which these DEGs are involved, KEGG database was used to analyze the pathway annotation of unigenes. The DEGs which had a KO ID could be categorized into pathway items. Among these, the top 20 pathways were significantly enriched with the standard p-value < 0.05 for down- and up-regulated DEGs (Fig. 2). The three most significantly enriched pathway items were ‘carbon fixation in photosynthetic organisms’, followed by ‘glyoxylate and dicarboxylate metabolism’ and ‘vitamin B6 metabolism’ for down-regulated; while ‘mismatch repair’, ‘thiamine metabolism’ and ‘arachidonic acid metabolism’ for up-regulated DEGs.
Pathway enrichment analysis of DEGs. a Top 20 KEGG terms enriched for down-regulated DEGs. b Top 20 KEGG terms enriched for up-regulated DEGs. The size of each circle represents the number of significantly differentially expressed genes enriched in the corresponding term. The rich factor was calculated using the number of enriched genes divided by the total number of background genes in the corresponding pathway. The p-value was calculated using the Benjamini-Hochberg correction. A term with P < 0.05 is considered significantly enriched
Expression of phytohormone-related genes in wheat plants in response to WDV infection
To investigate the involvement of phytohormone in wheat plants in response to WDV infection, we analyzed the gene expression profiling involved in pathways of biosynthesis and response of AUX, GAs, CKs, ABA, ET, and BRs. As shown in Fig. 3a, the related genes were mainly involved in categories corresponding to auxin response (GO:0009733), gibberellin biosynthetic process (GO:0009686), gibberellin metabolic process (GO:0009685), abscisic acid response (GO:0009737), cytokinin response (GO:0009735), ethylene response (GO:0009723), cellular response to ethylene stimulus (GO:0071369) and hormone response (GO:0009725). The main genes involved in biosynthesis and signaling transduction are listed in Table 2. Three genes, encoding auxin-induced protein 15A (SAUR), involved in the AUX signaling pathway, were consistently down-regulated. In the BRs mediated signaling pathway, the gene related with probable serine/threonine-protein kinase BSK3 (BSK) was decreased, while the gene encoding probable BRI1 kinase inhibitor 1 (BKI1) was increased. Meanwhile, the gene encoding steroid 5-alpha-reductase DET2 (Det2) involved in BRs biosynthesis, was down-regulated. The genes associated with probable protein phosphatase 2C 25 (PP2C) were induced in the ABA signaling pathway. The expression of genes encoding two-component response regulator ORR22 (B-ARR) involved in cytokinin-activated signaling pathway were up-regulated. However, genes related with two-component response regulator ORR4 (A-ARR) were down-regulated. Expression of the gene encoding protein ETHYLENE-INSENSITIVE 2 (EIN2) was decreased in ethylene-activated signaling pathway. In summary, most of the genes involved in signaling and biosynthesis pathways of AUX and BRs showed down-regulated expression, while genes involved in ABA, ET and CKs signaling pathways had up-regulated expression trends.
Gene ontology (GO) analysis of differentially expressed genes related to metabolism and signaling pathways of phytohormone and photosynthesis in response to wheat dwarf virus (WDV) infection in wheat (Triticum aestivum). a Gene ontology (GO) analysis of differentially expressed genes related to phytohormone metabolism and signaling pathways. b Gene ontology (GO) analysis of differentially expressed genes related to photosynthesis related pathways
Expression of genes involved in photosynthesis and chlorophyll metabolism
Previous studies revealed that virus infection can modify photosynthesis, and disturb chloroplast components and functions (Kyselakova et al. 2011; Manfre et al. 2011; Krenz et al. 2012). To elucidate the effect of WDV infection on chloroplast, we analyzed the expression pattern of the DEGs related to photosynthesis and chloroplast. As shown in Fig. 3b, the related DEGs were mainly involved in categories corresponding to chloroplast (GO:0009507), chloroplast part (GO:0044434), thylakoid (GO:0009579), chloroplast stroma (GO:0009570) and chloroplast thylakoid (GO:0009534). The main genes involved in the carbon fixation in photosynthetic organisms, photosynthesis, photosynthesis-antenna proteins and porphyrin and chlorophyll metabolism are listed in Table 3. Thirty eight genes, encoding ribulose bisphosphate carboxylase small chain (RBCS), probable ribose-5-phosphate isomerase 3 (RPI3), glyceraldehyde-3-phosphate dehydrogenase B (GAPDH), fructose-bisphosphate aldolase (FBA), fructose-1,6-bisphosphatas (FBP), phosphoglycerate kinase (PGK) and sedoheptulose-1,7-bisphosphatase (SPBase), involved in the carbon fixation in photosynthetic organisms, were down-regulated consistently. The DEGs in photosynthesis were also observed to be down-regulated consistently, including the genes encoding photosystem II repair protein PSB27-H1 (Psb27), ferredoxin (PetF), photosystem II 22 kDa protein (PsbS), oxygen-evolving enhancer protein 2 (PsbP), photosynthetic NDH subunit of lumenal location 3 (PsbQ) and photosystem II reaction center PSB28 protein (Psb28). In the photosynthesis-antenna proteins, the genes encoding photosystem I chlorophyll a/b-binding protein 6 (Lhca2), chlorophyll a-b binding protein of LHCII type III (Lhcb3), chlorophyll a-b binding protein CP24 (Lhcb6) and the putative chlorophyll (ide) b reductase NYC1 (NYC1) were consistently and greatly decreased. Expression of the gene encoding protein glutamyl-tRNA reductase 2 (HEMA2) was down-regulated in porphyrin and chlorophyll metabolism.
Validation of transcriptomics data by RT-qPCR
To quantify the transcriptome data, 10 DEGs were selected for RT-qPCR, including 4 up-regulated DEGs, [PP2Cc (ABA Pathway), BKI1 (BR Pathway), Rboh and RPM1 (plant-pathogen interaction)], and 6 down-regulated genes, [SAUR (AUX Pathway), RR5 (CK Pathway) and rbcS, FBA11, SBPase and RPI2 (carbon fixation in photosynthetic organisms)] (Fig. 4). The RNA samples from mock and WDV-infected plants for RNA-seq were also used for RT-qPCR. The trends in transcriptional variation by RT-qPCR for all selected genes were consistent with those of RNA-Seq, indicating that RNA-Seq is a reliable method to identify and select the differentially expressed genes in wheat in response to WDV infection.
Discussion
This study is, to our knowledge, the first large scale transcriptome RNA-Seq analysis of wheat upon WDV infection. It is also the first report on the changes of photosynthesis and phytohormone metabolism and signaling pathways in wheat responding to WDV infection.
Metabolic and signaling pathway of phytohormone may account for symptom formation
Changes in hormone metabolism and signaling are usually related with symptoms caused by virus infection and may lead to the hormone imbalance (Luis et al. 2019). As a result of this imbalance, plant height decreases and tiller number increases in WDV infected wheat plants. The majority of plant hormones are small and relatively simple molecules which are produced by specific organs and subsequently conveyed to target tissues to elicit a certain physiological response (Davies 1995). Thus, we analyzed the phytohormone-related DEGs following the pathway for each hormone.
AUX pathway
The term “auxin” originating from the Greek word “auxein” means “to grow”. A study revealed that RDV can reprogram a key step in auxin signaling cascades, resulting in enhanced viral infection and symptom development (Jin et al. 2016). In our study, the expression of DEGs encoding auxin-induced protein 15A and auxin-responsive protein SAUR72 that belong to SAUR family were down-regulated after WDV infection. SAUR family is one of the important gene families involved in auxin signaling-regulated plant growth and development. The plant growth processes in which SAUR family genes are involved include cell elongation (Chae et al. 2012), cell expansion (Spartz et al. 2012), light signaling (Roig-Villanova et al. 2007) and interactions with BRs (Favero et al. 2017), GA (Stamm and Kumar 2013) and ET (Li et al. 2015) in Arabidopsis. Our results suggested that reduced levels of SAUR in infected wheat plants may disturb plant growth processes, contributing to the stunting symptom development. Furthermore, excess tillering may be regulated through a very complex interplay among several hormones, including AUX and CKs.
CKs pathway
Previous study showed that cytokinins-deficient plants develop stunted shoots with smaller apical meristems, but the overproduction of cytokinins result in decreased susceptibility to virus infection (Werner et al. 2001; Pogany et al. 2004). In addition, biologically active cytokinin free bases and ribosides tend to become the inactive 9-glucosides after virus infection (Dermastia et al. 1995; Suttle and Mornet 2005). Interestingly, we found that the expression of genes related with B-ARR and A-ARR were up-regulated and down-regulated, respectively. Mutations in B-ARR, key regulators of primary cytokinin response genes, result in reduced shoot regeneration (Ishida et al. 2008). By contrast, overexpressing A-ARR encoding negative regulators of cytokinin signaling inhibits shoot formation (Buechel et al. 2010). Thus, we speculate that the infection of WDV may result in the imbalance of CKs leading to an abnormal plant growth.
BRs pathways
Brassinosteroids, another class of growth-promoting regulator, regulate many aspects of plant growth and development including cell expansion and elongation (in association with auxin), and vascular differentiation (Li and Chory 1997). The necessity of brassinosteroids in plant growth and development was proved by identification of many BR-deficient dwarf mutants and subsequent BRs treating experiments (Altmann 1998a, b; Altmann 1999; Bishop and Yokota 2001). The brassinosteroid-signaling kinases (BSKs) are critical in BRs signal transduction (Li et al. 2019). BSK3 is the only BSK member involved in BR-mediated plant root growth (Ren et al. 2019). Moreover, Det2 (which encodes a steroid 5α-reductase that catalyzes the formation of campestanol with campesterol as substrates) is also involved in BRs biosynthesis (Lv et al. 2018). BRI1 kinase inhibitor1 (BKI1) negatively regulates BRI1 by preventing BRI1 from combining with BRI1-associated kinase (BAK1) to form an active receptor complex in the BRs signaling pathway (Jiang et al. 2015). The decreased expression of DEGs encoding BSK3 and Det2, and increased BKI1 in our study suggested that WDV infection may impair BRs biosynthesis and signaling pathway in wheat plant, and subsequently block its normal response to BRs.
Other pathways
Other phytohormones (e.g., ET and ABA) are also involved in the growth and development of plants. ET plays an active roles in disease resistance, but ABA acts as a negative regulator of disease resistance (Mauch-Mani and Mauch 2005; Agudelo-Romero et al. 2008; Pallas and Garcia 2011; Atkinson and Urwin 2012). In our study, decreased expression of gene encoding EIN2 in ET signaling pathway suggests that the ET signal transduction pathway might be blocked to some extent in WDV infected plants. At the same time, the up-regulated gene related with PP2C indicates that ABA signaling pathway might be induced in WDV infected plants.
Disordered photosynthesis is another factor responsible for symptom formation
The virus adjusts multiplex processes, including sugar effluence, carbon distribution and phloem conveyance of metabolites inside the host cells (Balachandran et al. 1995; Olesinski et al. 1995; Chen et al. 2010). In our study, the expression levels of DEGs encoding RBCS, RPI3, GAPDH, FBA, FBP, PGK and SPBase in the pathway of carbon fixation, PetF, Psb27, PsbS, PsbP, PsbQ and Psb28 in photosynthesis and HEMA2 in porphyrin and chlorophyll metabolism were all down-regulated after WDV infection. Thus, since chlorophyll is a key compound in green plants, this could explain, to some extent, the reason of yellowing induced due to WDV infection. Regarding the pathway of the photosynthesis-antenna proteins, four genes (encoding Lhca2, Lhcb3, Lhcb6 and NYC1) were down-regulated. These findings indicate that WDV infection may lead to decreased photoelectron capture and photosynthetic ability in wheat leaves after infection. Consequently, this might affect the enzymes involved in photosynthesis, alerting the structure and function of chloroplast during WDV infection. Thus, the up-regulation and down-regulation of the genes mentioned above caused by WDV infection might be the key elements contributing to the yellowing symptoms development observed in infected plants.
Conclusions
Based on the current studies, it is clear that WDV infection intensively impacts many aspects of the physiology of wheat plants, including metabolism and signaling of phytohormone and photosynthetic capacity. We highlight the link between metabolic and signaling pathway related to phytohormones and photosynthesis in wheat and symptoms development upon WDV infection. Our results suggest that WDV infection may have significant effect on signaling and biosynthesis of AUX, CKs, BRs and ET, and meanwhile contribute to symptom development through reduced expression of related gene excepting CKs pathway. Additionally, the ABA-mediated signaling pathway negatively regulates the defense system during WDV infection. A compilation of the genes related to phytohormones and photosynthesis pathway would provide an ideal list of candidates to investigate the interaction between WDV and host plants. Our study provides valuable and novel information to understand the development of viral symptoms.
Methods
Virus, insect vector, plant materials and inoculation process
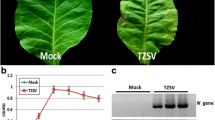
An isolate of WDV (wheat strain) collected from Hancheng, Shaanxi in 2015 (accession number: JQ647498.1) was used in this study. Viruliferous and healthy leafhoppers (P. alienus) were reared separately on wheat plants (cv. Yangmai 12) using insect-proof containment chambers at a temperature of 22 °C and a photoperiod of 16/8 h (day/night) (Wang et al. 2019b). A highly susceptible wheat cultivar, Yangmai 12 was used for the experiment. The wheat plants were grown in pots that were kept in an illumination incubator at 22 °C with a 16 h photoperiod. For WDV inoculation, leafhoppers were fed on WDV infected plant for 4 days and then separately transferred to healthy seedlings at the single-leaf stage (3 insects per plant). The treatment with healthy leafhopper was used as negative control. All the leafhoppers were removed at 4 days post-inoculation (dpi). At 20 dpi, WDV-infected samples with typical dwarfing and yellowing symptoms (named W20–1, W20–2 and W20–3) and mock-treated samples (named CK-1, CK-2 and CK-3) were harvested (Fig. 5). Systemic viral infection was subsequently determined by PCR analysis as described previously (Wang et al. 2014).
Preparation of libraries for RNA-Seq
To minimize the bias of random changes from a single sample, wheat top leaves were sampled from three independent treatments and then total RNA of W20–1, W20–2, W20–3 and CK-1, CK-2, CK-3 were pooled equally for further experiments, respectively. Total RNA from top leaves was isolated using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Agilent 2100 Bioanalyzer (plant RNA Nano Chip, Agilent, USA) was used to determine the quality and concentration of RNA. The library construction and sequencing via Illumina HiSeq™ 2000 were performed at the Novogene (Novogene Bioinformatics Technology Co. Ltd., Beijing, China), and approximately 100 bp paired-end (PE) raw reads were generated.
Data filtering, de novo assembly
Raw reads were produced via a series of processes, along with dirty reads which would negatively affect subsequent bioinformatics analysis. After filtering of adaptor sequences and low quality reads (the rate of reads with quality value < = 3 is more than 50% or the percentage of unknown sequences ‘N’ is more than 3%), the cleaned reads were assembled into unigenes by Trinity (Grabherr et al. 2011). Longer fragments and contigs were formed via a certain length of overlap (250–300 bp). The contigs which cannot be extended on both ends were defined as unigenes.
Mapping and calculation of RPKM
The assembled transcriptome data was used as reference sequences. All the clean reads from W20 group and CK group were separately mapped back to the reference sequences by HISAT2 (http://ccb.jhu.edu/software/hisat2/faq.shtml). RPKM (Reads Per Kilobase of exon Million mapped sequence reads) was used to calculate the expression level of unigene and the formula was as follows:
The method of DESeq2 (Anders and Huber 2010) was used to screen genes which were differentially expressed between the treatment group of mock and WDV- inoculated plants.
Gene annotation and gene ontology (GO) enrichment analysis
Although the sketch of common wheat genome is nearly completed, the functions of a large number of wheat genes are still unknown owing to the hugeness and complexity of its genome. Functional unigene annotation was mainly proceeded under the reference of model plant: Brachypodium distachyon. All the DEGs were firstly blasted to B. distachyon database by HISAT2, retrieving genes with the highest sequence similarity with the DEGs. To classify the gene function of unigenes, gene ontology (GO) enrichment analysis was performed. All DEGs were firstly mapped to GO terms in the database (http://www.geneontology.org) and then the GO enrichment analysis was carried out by Fisher test with Bonferroni correction. Taking corrected p-value < 0.01 as a threshold, GO terms fulfilling this condition were defined as significantly enriched GO terms in DEGs.
Pathway analysis for DEGs
KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa and Goto 2000) was used to analyze the complex biological process of DEGs. We firstly obtained all the pathway items in which all the DEGs are involved, and then calculated the p-value using Fisher test based on hypergeometric distribution to screen the significantly enriched pathway items, taking corrected p-value < 0.05 as threshold.
Validation by reverse transcription quantitative PCR (RT-qPCR)
Expression level of all the selected genes was assayed by SYBR Green RT-qPCR. We extracted RNA from both infected and healthy wheat samples and then used 1 μg RNA of each sample to synthesize cDNA by reverse transcription using TRUEscript 1st Strand cDNA Synthesis Kit (Aidlab). RT-qPCR was performed using the TransStart® Green qPCR SuperMix (Transgen) with the ABI 7500 Real Time PCR system (Applied Biosystems). The PCR reactions were subjected to an initial denaturation step of 94 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The β-tubulin gene (accession: U76896) was used as an internal control for studying the expression level of target genes. Relative gene expression levels were analyzed with 2-∆∆CT method (Livak and Schmittgen 2001). All reactions were performed in three technical and biological replicates. The primers for reference and target genes were designed respectively according to wheat sequences from GenBank and the unigene sequences. The detailed information of primers showed in Additional file 1: Table S1.
Availability of data and materials
Not applicable.
Abbreviations
- A-ARR:
-
Two-component response regulator ORR4
- ABA:
-
Abscisic acid
- AUX:
-
Auxin
- B-ARR:
-
Two-component response regulator ORR22
- BKI1:
-
Probable BRI1 kinase inhibitor 1
- BRs:
-
Brassinosteroids
- BSK:
-
Probable serine/threonine-protein kinase BSK3
- CKs:
-
Cytokinins
- DEGs:
-
Differentially expressed genes
- Det2:
-
Steroid 5-alpha-reductase DET2
- EIN2:
-
Protein ETHYLENE-INSENSITIVE 2
- ET:
-
Ethylene
- FBA:
-
Fructose-bisphosphate aldolase
- FBP:
-
Fructose-1,6-bisphosphatase
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase B
- GAs:
-
Gibberellins
- GO:
-
Gene ontology
- HEMA2:
-
Glutamyl-tRNA reductase 2
- JA:
-
Jasmonic acid
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- Lhca2:
-
Photosystem I chlorophyll a/b-binding protein 6
- Lhcb3:
-
Chlorophyll a-b binding protein of LHCII type III
- Lhcb6:
-
Chlorophyll a-b binding protein CP24
- NYC1:
-
Probable chlorophyll (ide) b reductase NYC1
- PetF:
-
Ferredoxin
- PGK:
-
Phosphoglycerate kinase
- PP2C:
-
Probable protein phosphatase 2C 50
- Psb27:
-
Photosystem II repair protein PSB27-H1
- Psb28:
-
Photosystem II reaction center PSB28 protein
- PsbP:
-
Oxygen-evolving enhancer protein 2
- PsbQ:
-
Photosynthetic NDH subunit of lumenal location 3
- PsbS:
-
Photosystem II 22 kDa protein
- RBCS:
-
Ribulose bisphosphate carboxylase small chain
- RDV:
-
Rice dwarf virus
- RPI3:
-
Probable ribose-5-phosphate isomerase 3
- SA:
-
Salicylic acid
- SAUR:
-
Auxin-induced protein 15A
- SPBase:
-
Sedoheptulose-1,7-bisphosphatase
References
Adolph KW, Haselkorn R. Photosynthesis and the development of blue-green algal virus N-1. Virology. 1972;47:370–4.
Agudelo-Romero P, Carbonell P, de la Iglesia F, Carrera J, Rodrigo G, Jaramillo A, et al. Changes in the gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus. Virol J. 2008;5:92.
Alazem M, Lin NS. Roles of plant hormones in the regulation of host-virus interactions. Mol Plant Pathol. 2015;16:529–40.
Allie F, Pierce EJ, Okoniewski MJ, Rey C. Transcriptional analysis of South African cassava mosaic virus-infected susceptible and tolerant landraces of cassava highlights differences in resistance, basal defense and cell wall associated genes during infection. BMC Genomics. 2014;15:1006.
Altmann T. Recent advances in brassinosteroid molecular genetics. Curr Opin Plant Biol. 1998a;1:378–83.
Altmann T. A tale of dwarfs and drugs: brassinosteroids to the rescue. Trends Genet. 1998b;14:490–5.
Altmann T. Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta. 1999;208:1–11.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Atkinson NJ, Urwin PE. The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot. 2012;63:3523–43.
Balachandran S, Hull RJ, Vaadia Y, Wolf S, Lucas WJ. Alteration in carbon partitioning induced by the movement protein of tobacco mosaic virus originates in the mesophyll and is independent of change in the plasmodesmal size exclusion limit. Plant Cell Environ. 1995;18:1301–10.
Balachandran S, Hurry VM, Kelley SE, Osmond CB, Robinson SA, Rohozinski J, et al. Concepts of plant biotic stress. Some insights into the stress physiology of virus-infected plants, from the perspective of photosynthesis. Physiol Plant. 1997;100:203–13.
Berens M, Berry H, Mine A, Argueso C, Tsuda K. Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol. 2017;55:401–25.
Buechel S, Leibfried A, To JPC, Zhao Z, Andersen SU, Kieber JJ, et al. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur J Cell Biol. 2010;89:279–84.
Bishop GJ, Yokota T. Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 2001;42:114–20.
Bisztray G, Gaborjanyi R, Vacke J. Isolation and characterisation of wheat dwarf virus found for the first time in Hungary. J Plant Dis Prot. 1989;96:449–54.
Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, et al. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71:684–97.
Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–32.
Commandeur U, Huth W. Differentiation of strains of wheat dwarf virus in infected wheat and barley plants by means of polymerase chain reaction. J Plant Dis Prot. 1999;106:550–2.
Davies PJ. Plant hormones: physiology, biochemistry and molecular biology. 2nd ed. Dordrecht: Kluwer Academic; 1995.
De Vleesschauwer D, Xu J, Hofte M. Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front Plant Sci. 2014;5:1–15.
Dermastia M, Ravnikar M, Kovac M. Increased cytokinin-9-glucosylation in roots of susceptible Solanum tuberosum cultivar infected by potato-virus YNTN. Mol Plant-Microbe Interact. 1995;8:327–30.
Díaz-Vivancos P, Clemente-Moreno MJ, Rubio M, Olmos E, Garcia JA, Martínez-Gómez P, et al. Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. J Exp Bot. 2008;59:2147–60.
Favero DS, Le KN, Neff MM. Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J. 2017;89:1133–45.
Fraser RSS, Whenham RJ. Plant growth regulators and virus infection: a critical review. Plant Growth Regul. 1982;1:37–59.
Funayama-Noguchi S, Terashima I. Effects of Eupatorium yellow vein virus infection on photosynthetic rate, chlorophyll content and chloroplast structure in leaves of Eupatorium makinoi during leaf development. Funct Plant Biol. 2006;33:165–75.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Guo DP, Guo YP, Zhao JP, Liu H, Peng Y, Wang QM, et al. Photosynthetic rate and chlorophyll fluorescence in leaves of stem mustard (Brassica juncea var. tsatsai) after turnip mosaic virus infection. Plant Sci. 2005;168:57–63.
Gutierrez C. Geminivirus DNA replication. Cell Mol Life Sci. 1999;56:313–29.
Hanssen IM, van Esse HP, Ballester AR, Hogewoning SW, Parra NO, Paeleman A, et al. Differential tomato transcriptomic responses induced by Pepino mosaic virus isolates with differential aggressiveness. Plant Physiol. 2011;156:301–18.
He Y, Zhang H, Sun Z, Li J, Hong G, Zhu Q, et al. Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 2017;214:388–99.
Ishida K, Yamashino T, Yokoyama A, Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57.
Jameson PE, Clarke SF. Hormone-virus interactions in plants. Crit Rev Plant Sci. 2002;21:205–28.
Jia MA, Li YQ, Lei L, Di DP, Miao HQ, Fan ZF. Alteration of gene expression profile in maize infected with a double-stranded RNA fijivirus associated with symptom development. Mol Plant Pathol. 2012;13:251–62.
Jiang JJ, Wang T, Wu ZW, Wang J, Zhang C, Wang HJ, et al. The intrinsically disordered protein BKI1 is essential for inhibiting BRI1 signaling in plants. Mol Plant. 2015;8:1675–8.
Jin L, Qin QQ, Wang Y, Pu YY, Liu LF, Wen X, et al. Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 2016;12(9):e1005847.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Koornneef A, Pieterse CMJ. Cross talk in defense signaling. Plant Physiol. 2008;146:839–44.
Krenz B, Jeske H, Kleinow T. The induction of stromule formation by a plant DNA-virus in epidermal leaf tissues suggests a novel intra- and intercellular macromolecular trafficking route. Front Plant Sci. 2012;3:291.
Kundu J, Gadiou S, Cervena G. Discrimination and genetic diversity of Wheat dwarf virus in the Czech Republic. Virus Genes. 2009;38:468–74.
Kyselakova H, Prokopova J, Naus J, Novak O, Navratil M, Safarova D, et al. Photosynthetic alterations of pea leaves infected systemically by pea enation mosaic virus: a coordinated decrease in efficiencies of CO2 assimilation and photosystem II photochemistry. Plant Physiol Bioch. 2011;49:1279–89.
Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–38.
Li Z, Shen J, Liang J. Genome-wide identification, expression profile, and alternative splicing analysis of the brassinosteroid-signaling kinase (BSK) family genes in Arabidopsis. Int J Mol Sci. 2019;20:1138.
Li ZG, Chen HW, Li QT, Tao JJ, Bian XH, Ma B, et al. Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci Rep. 2015;5:12477.
Lindblad M, Sigvald R. Temporal spread of wheat dwarf virus and mature plant resistance in winter wheat. Crop Prot. 2004;23:229–34.
Lindsten K, Lindste B. Wheat dwarf-an old disease with new outbreaks in Sweden. J Plant Dis Prot. 1999;106:325–32.
Liu Y, Jin W, Wang L, Wang XF. Replication-associated proteins encoded by Wheat dwarf virus act as RNA silencing suppressors. Virus Res. 2014;190:34–9.
Liu Y, Wang B, Vida G, Cseplo-Karolyi M, Wu BL, Wu YH, et al. Genomic analysis of the natural population of Wheat dwarf virus in wheat from China and Hungary. J Integr Agr. 2012;11:2020–7.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–8.
Lu J, Du ZX, Kong J, Chen LN, Qiu YH, Li GF, et al. Transcriptome analysis of Nicotiana tabacum infected by Cucumber mosaic virus during systemic symptom development. PLoS One. 2012;7:e43447.
Luis A, Sofía M, Ondrej N, Regina F, Analía D, María F, et al. Mal de Río Cuarto virus infection causes hormone imbalance and sugar accumulation in wheat leaves. Plant Biol. 2019;19:112–28.
Lv BS, Tian HY, Zhan F, Liu JJ, Lu SC, Bai MY, et al. Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 2018;14(1):e1007144.
Manfre A, Glenn M, Nunez A, Moreau RA, Dardick C. Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid–independent mechanism. Mol Plant-Microbe Interact. 2011;24:315–27.
Mauch-Mani B, Mauch F. The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol. 2005;8:409–14.
Mochizuki T, Ogata Y, Hirata Y, Ohki ST. Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with Cucumber mosaic virus. Mol Plant Pathol. 2014;15:242–54.
Najar A, Makkouk KM, Boudhir H, Kumari SG, Zarouk R, Bessai R, et al. Viral diseases of cultivated legume and cereal crops in Tunisia. Phytopathol Mediterr. 2000;39:423–32.
Olesinski AA, Lucas WJ, Galun E, Wolf S. Pleiotropic effects of tobacco mosaic virus movement protein on carbon metabolism in transgenic tobacco plants. Planta. 1995;197:118–26.
Pallas V, Garcia JA. How do plant viruses induce disease? Interactions and interference with host components. J Gen Virol. 2011;92:2691–705.
Pineda M, Sajnani C, Baron M. Changes induced by the Pepper mild mottle tobamovirus on the chloroplast proteome of Nicotiana benthamiana. Photosynth Res. 2010;103:31–45.
Pogany M, Koehl J, Heiser I, Elstner EF, Barna B. Juvenility of tobacco induced by cytokinin gene introduction decreases susceptibility to Tobacco necrosis virus and confers tolerance to oxidative stress. Physiol Mol Plant Pathol. 2004;65:39–47.
Ren H, Willige BC, Jaillais Y, Geng S, Park MY, Gray WM, et al. BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 2019;15:e1007904.
Rizza S, Conesa A, Juarez J, Catara A, Navarro L, Duran-Vila N, et al. Microarray analysis of Etrog citron (Citrus medica L.) reveals changes in chloroplast, cell wall, peroxidase and symporter activities in response to viroid infection. Mol Plant Pathol. 2012;13:852–64.
Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, et al. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26:4756–67.
Sanchez G, Gerhardt N, Siciliano F, Vojnov A, Malcuit I, Marano MR. Salicylic acid is involved in the Nb-mediated defense responses to Potato virus X in Solanum tuberosum. Mol Plant-Microbe Interact. 2010;23:394–405.
Song XS, Wang YJ, Mao WH, Shi K, Zhou YH, Nogues S, et al. Effects of cucumber mosaic virus infection on electron transport and antioxidant system in chloroplasts and mitochondria of cucumber and tomato leaves. Physiol Plant. 2009;135:246–57.
Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, et al. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70:978–90.
Stamm P, Kumar PP. Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 2013;32:759–69.
Suttle JC, Mornet R. Mechanism-based irreversible inhibitors of cytokinin dehydrogenase. J Plant Physiol. 2005;162:1189–96.
Vacke J. Wheat dwarf virus disease. Biol Plant. 1961;3:228–33.
Wang H, Liu Y, Zhang L, Kundu JK, Liu W, Wang X. ADP ribosylation factor 1 facilitates spread of wheat dwarf virus in its insect vector. Cell Microbiol. 2019a;21:e13047.
Wang H, Wu N, Liu Y, Kundu JK, Liu W, Wang X. Higher bacterial diversity of gut microbiota in different natural populations of leafhopper vector does not influence WDV transmission. Front Microbiol. 2019b;10:1144.
Wang Y, Mao Q, Liu W, Mar TT, Wei T, Liu Y, et al. Localization and distribution of Wheat dwarf virus in its vector leafhopper, Psammotettix alienus. Phytopathology. 2014;104(8):897–904.
Werner T, Motyka V, Strnad M, Schmulling T. Regulation of plant growth by cytokinin. Proc Natl Acad Sci U S A. 2001;98:10487–92.
Weyers JDB, Paterson NW. Plant hormones and the control of physiological processes. New Phytol. 2001;152:375–407.
Wu LJ, Wang SX, Chen X, Wang XT, Wu LC, Zu XF, et al. Proteomic and phytohormone analysis of the response of maize (Zea mays L.) seedlings to sugarcane mosaic virus. PLoS One. 2013;8:e70295.
Xie J, Wang X, Liu Y, Peng Y, Zhou G. First report of the occurrence of Wheat dwarf virus in wheat in China. Plant Dis. 2007;91:111.
Zhao J, Zhang X, Hong Y, Liu Y. Chloroplast in plant-virus interaction. Front Microbiol. 2016;7:1565.
Zhou Y, Xu Z, Duan C, Chen Y, Meng Q, Wu J, et al. Dual transcriptome analysis reveals insights into the response to Rice black-streaked dwarf virus in maize. J Exp Bot. 2016;67(15):4593–609.
Zhu S, Gao F, Cao X, Chen M, Ye G, Wei C, et al. The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 2005;139(4):1935–45.
Acknowledgments
We thank Dr. Hazen BE (Willows End scientific editing and writing, USA) for critically reading and revising the manuscript.
Funding
This research was supported by the Funds for International Cooperation and Exchange of the National Natural Science Foundation (31861133020).
Author information
Authors and Affiliations
Contributions
XW and LL conceived and designed the experiments; YL, CS, YL and LL carried out the experiments and data analysis; YL and XW drafted the manuscript; XW and CS discussed the results and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary information
Additional file 1: Table S1.
Primers for RT-qPCR.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, Y., Liu, Y., Spetz, C. et al. Comparative transcriptome analysis in Triticum aestivum infecting wheat dwarf virus reveals the effects of viral infection on phytohormone and photosynthesis metabolism pathways. Phytopathol Res 2, 3 (2020). https://doi.org/10.1186/s42483-019-0042-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-019-0042-6