Abstract
Background
Actinomycetes are prolific producers of bioactive compounds which can be used to combat microbial infections. Microbial infections have resulted to increase in mortality and morbidity rates world-wide, especially in developing nations. This study was carried out to isolate and identify actinomycetes with potentials of synthesizing bioactive compounds, to produce and partially purify antimicrobial extracts, to characterize bioactive compounds in the extracts, to assess their antibacterial activity and to profile other biological properties of the bioactive compounds synthesized by the actinomycetes.
Results
Streptomyces sp. SUI (MT584797), Streptomyces sp. SW72IV (MT584818) and Streptomyces sp. SW72VII (MT584816) were isolated and identified. Ultraviolet–visible spectra absorption ranged between 241 and 251 nm wavelengths showing the polyene or chromone and unsaturation nature of the natural compounds. Infrared (IR) spectra indicated functional groups such as hydroxyl, aliphatic bromo, carbonyls, esters, carboxylic acids and silicon oxy compounds in the compounds produced by the three strains. Gas Chromatography-Mass Spectrum (GC–MS) identified elaidic acid isopropyl ester (32.11%), Octadec-9-enoic acid (17.44%) and 2, 3-dihydroxyl elaidate (10%) to be mostly produced by Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII respectively. The three strains exhibited antimicrobial activity against Bacillus sp. Pseudomonas aeruginosa ATCC 9077, Staphylococcus aureus ATCC 700699, Candida albicans and Aspergilus flavus.
Conclusion
The results showed that the three strains of Streptomyces could be sources of antimicrobial bioactive compounds and other secondary metabolites that can be used in the production of pharmaceutical bioactive agents that are effective against pathogens, and production of biological materials that can be used in cosmetics and food industries.
Similar content being viewed by others
Background
Actinobacteria are a diverse group of Gram positive filamentous bacteria. The members of this group consist of most important microflora found in most places such as soil, fresh water, marine, lakes as well as dumping sites. Extreme and unexplored areas in the environment could be home to antimicrobial sources (Qinn et al. 2020). They are characteristically known to produce distinctive odor from a chemical compound called geosmin usually perceived when soil is tilled. Actinomycetes play important roles in recycling of organic materials in the environment (Malibari 1991).
The most studied genus has been Streptomyces for its bioactive compounds. They are endowed with secondary metabolites that are bioactive in nature. Most of these metabolites are antimicrobials, enzymes, enzyme inhibitor, antioxidants, anticancer and so on (Barka et al. 2016; Sharma and Thakur 2020). They are prominent producers of so many natural products that have found applications in pharmaceutical, food and chemical industries (Berdy 2005). Natural products obtained from actinobacteria contain bioactive compounds that are diverse in structures and functions. Most drugs primarily originated from these actinobacteria and are active against many emerged and re-emerging pathogens (Tripathi et al. 2004; Singh et al. 2012). Most known antimicrobial drugs applied in agriculture and medicines have been sourced from natural bioactive compounds synthesized by actinobacteria (Sanglier et al. 1996). About seven thousand compounds documented in the compendium of natural products are from the actinobacteria (Berdy 2005). Hence, their ability to produce these compounds is very essential to pharmaceutical and chemical industries (Berdy 2005). It is because of this that the research efforts at sourcing for novel strains of actinobacteria that possess the ability to synthesize bioactive compounds have been intensified especially in recent times (Singh et al. 2008). The last few years have witnessed a surge in the number of Streptomyces isolated and identified (Nandhini et al. 2015). Abd-Ellatif et al. (2019) isolated a marine strain that was capable of producing linoleic acid. Linoleic acids and its derivatives have useful applications in the treatment of cancer, depression and high blood pressure. Pheromone for instance has been identified among metabolites synthesized by Streptomyces lavendulocolor UHB-9 that has the potential of producing cis-hecadecenal. Cis-hexadecenal is an antimicrobial and a pheromone. It is a world-wide research activity to search for strains of actinobacteria with the potential to produce secondary metabolites with active biological properties that can have wide applications in pharmaceutical, industrial and agricultural sectors (Siddharth et al. 2020).
Resistance to antibiotics is spreading rapidly and this has attracted the attention of researchers to shift their focus to the discovery of novel and potent bioactive compounds of actinobacteria origin that can be used to treat infections caused by microbes (Sebak et al. 2021). Microbial infection is responsible for high mortality and morbidity rates especially in poor and developing countries of the world. According to Centre for Disease and Control, (CDC), resistance to antimicrobial drugs is responsible for nearly a million deaths annually (CDC 2019). It has debilitating effects on the economic power of patients and generally lowers productivity generally (Siddharth et al. 2020).
The objectives of this study were to isolate and identify Actinobacteria isolates from Lake Gerio, Yola, Adamawa State, Nigeria, to characterize secondary metabolites synthesized by the Actinobacteria and to assess their antibacterial activity as well as to profile the biological properties of different secondary metabolites produced by the identified strains.
Methods
Sampling location
Lake Gerio is located within 9° 16′N–9° 21′N and 12° 24′E–12° 27′E of Yola, Adamawa State, North eastern part of Nigeria. It was naturally formed from cut off creek of River Benue that runs in between Girei and Yola North Local Government Area Councils of the State. It is about 2,500,000 m2 in size and it is mostly used for fishing and farm irrigation (Abiodun and Miller 2007). Water samples were collected in June 2019. The pH of the water sample collected was between 6.8 and 9.0 while the temperature was 33 °C.
Isolation of actinomycetes from collected Lake Gerio samples
Ten samples each of water were collected randomly from different points within Lake Gerio using sterile sampling bottles. The samples were immediately transported in a sterile container to the Industrial Biotechnology laboratory, Department of Biotechnology, Modibbo Adama University of Technology, Yola, Nigeria.
Actinobacteria were isolated using pour-plate technique after each sample was serially diluted according to previously described method of Harrigan and McCance (1976). 1.0 mL of serially was pour-plated in starch casein agar. Incubation was done at 28 ± 2 °C for 5–7 days. Different formed colonies were sub-cultured on starch casein agar and incubated for another 5–7 days at 28 ± 2 °C to obtain pure cultures which were preserved on starch casein agar slants at 4 °C in a refrigerator (Hisense). The pure cultures were used for further analyses.
Screening for antibacterial activity in isolated actinobacteria
Antibacterial activity of the isolated actinomycetes was assessed by perpendicular streaking method described by Oskay (2009). Screening for antibacterial activity was done on Mueller Hinton agar with P. aeruginosa ATCC 9077 and Bacillus sp. P. aeruginosa ATCC 9077 was obtained from Biological Science Department, Redeemers University, Ede, Osun State, Nigeria while Bacillus sp. was a local isolate from obtained from Microbiology department, Modibbo Adama University of Technology, Yola, Adamawa State, Nigeria. Those that exhibited inhibitory activity against the two test bacteria were selected for cultural, biochemical characteristics study and molecular identification using 16S rRNA gene.
Cultural and biochemical characteristics and molecular identification of the selected isolates
The cultural and morphological study of the three selected actinobacteria isolates SUI, SW72IV and SW72VII were done using the description in International Streptomyces Project with five ISP media, starch casein agar and nutrient agar (Shirling and Gottlieb 1966a, b; Küster and Williams 1964; Atta and Ahmad 2009) while pigment production was observed on three ISP media (Shirling and Gottlieb 1966a, b).
The abilities of the selected isolates to utilize different sugars and to produce enzymes were carried out using the methods described by Fawole and Oso (2004) and Collins et al. (1995). Sugars that included glucose (control), galactose, arabinose, glycerol, xylose, meso-inositol, rhamnose, D-mannitol starch, lactose, maltose, sucrose and fructose were used to test for sugar utilization. The potential of Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII to produce oxidase, catalase, urease, gelatinase was done using the method of Collins et al. (1995). Methyl red and Voges Proskaeur tests were carried with the method described by Prescott et al. (1991).
Molecular identification of the selected actinomycetes using 16S rRNA gene
Extraction of genomic DNA
Pure cultures of Streptomyces isolates SUI, SW72IV and SW72VII were grown in starch casein broth and incubated on a shaker at 250 rpm and at a temperature of 28 ± 2 °C for 5 days. The suspension was cold centrifuged at 10,000 rpm for 10 min. The mycelium was then used for genomic DNA extraction using Norgen DNA extraction kit (Model 24700 Norgen, Canada) according to the instructions of the manufacturer. The extracted DNA was purified and stored on ice pack for PCR amplification.
Identification of the actinomycetes isolates
PCR amplification
The 16S rRNA gene of actinomycetes isolates SUI, SW72IV and SW72VII were amplified using forward primer 243F 5′GGATGAGCCCGCGGCCTA3′ and reverse primer A3 5′CCAGCCCCACCTTCGAC-3′ (Monciardini et al. 2002). The PCR reaction was carried out with a final volume of 50 μL that contained 1.0 μL of template DNA, 10 mM Tris–HCl 1.5 U of Taq DNA polymerase (Applied Biosystem), 1.5 mM MgCl2, each primer was 500 nM while for each dNTP 0.2 mM was added (Inqaba Biotechnological, Pretoria, South Africa). The PCR reactions were carried out in PCR system (Applied Biosystem) with initial denaturation at 95 °C for five minutes. This was concluded with an extension at 72 °C with lasted for 10 min. The amplified product of 2 μL was confirmed on agarose gel electrophoresis of 1.0% (w/v) agarose powder dissolved in TBE buffer of pH 8.2 at 100 V for 90 min done using Bio Rad set. A molecular ladder of 100 bp (NEB’s fast DNA ladder) was added to serve as standard. After this, the gel was tracked with xylene cyanol FF and the gel was viewed under the illumination of UV light (Sanger and Coulson 1975).
Sequencing of amplified 16S rRNA gene and phylogenetic analysis
The amplified DNA was purified by PCR clean up and then it was sequenced in an automated DNA sequencer (BigDye Terminator Kit in a 3510 ABI Sequencer, Inqaba Biotechnological, Pretoria, South Africa) using the dideoxy chain termination method (Sanger and Coulson 1975). The 16S rRNA gene sequences of isolates SUI, SW72IV and SW72VII were analyzed and comparison with other deposited sequences was done using Entrez search engine at NCBI. The similarity between each isolate and typed Streptomyces strains at GenBank was done using BLAST program. Furthermore, multiple sequence alignment as well as the construction of the phylogenetic trees was done with molecular evolutionary genetics analysis (MEGA X) (Kumar et al. 2018). The gene sequences for Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII were deposited at National Center for Biotechnology Information (NCBI) and they were given accession numbers.
Production, extraction and partial purification of the bioactive compounds
Production of the secondary metabolites was carried out by submerged fermentation. A three days old seed inoculum of 100 mL was used to inoculate 5000 mL of sterile starch casein broth and was incubated at 28 ± 2 °C for 10 days. The fermentation was halted on the tenth day and the broth was filtered using Whatman No1 filter paper. The filtrate was then mixed with ammonium sulphate at 70% (w/v) to precipitate out proteinous materials. The resulted suspension was centrifuged at 10,000 rpm for 10 min.
Furthermore, the solvent extraction of the bioactive compounds was carried out by mixing equal volume of graded ethyl acetate and the filtrate in a separating funnel. The suspension was thoroughly mixed together for about 30 min and allowed to settle down for 20 min. The extract recovered from the ethyl acetate was partially purified using a column chromatography. The column was packed with solid phase silica gel of 100–200 mesh while ethyl acetate was used as mobile phase. 3.0 mL fractions were collected from the column till the introduced crude extract was completely eluted. Each fraction was tested for antibacterial activity and those that exhibited activity were pooled together and concentrated in rotatory evaporator at 60 °C.
Antimicrobial determination
Determination of antimicrobial activity of the actinobacteria strains was determined with Bacillus sp., P. aeruginosa ATCC 9077, S. aureus ATCC 700,699, C. albicans and A. flavus using agar well diffusion and disc diffusion methods (Wu 1984; Bauer et al. 1959). A well of 6 mm was made at the center of the petri dishes where the partially purified extract of 40 μL of 1 mg/mL concentration was introduced. Ethyl acetate (99.5%) was use as the negative control while both gentamicin (Oxoid, 10 μg) and fluconazole (Oxoid 25 μg) were used as positive controls. The diameter of the zone of inhibition was measured with a metric ruler and recorded in millimeter. All data were recorded in triplicates and mean values with standard deviation were determined.
Characterization of chemical compounds in the partially purified extracts
The UV–Vis, IR and GC–MS spectra
The UV–Vis spectra of the partially purified extracts were determined between the range 200–900 nm wavelengths using a UV-Jenway Model 6705 (California, USA) while the IR spectrum was determined with Buck Scientific Model 530 spectrophotometer (East Norwalk, USA) in scanned wavelengths that were between 4000 and 400 cm−1. Characterization of the bioactive compounds present in the extract was carried out with gas chromatography coupled to a mass spectrometer in Shidmazu GC 2010 (Japan) coupled with slit injection detector system as described by Yakubu et al. (2018). Briefly, 1.5 μL of the partially purified extract was injected into a capillary column of dimension while helium gas was used as carrier which flowed at the rate of 1.5 mL/min. The temperature of the column was initially set at 60 °C and lasted for 3 min. The temperature was raised to 250 °C and maintained throughout the running of the sample. The mass spectrometer was operated in the electron ionization mode at 70 eV while scanning was between 45 and 600 m/z. The identification of the compounds in the extract was done by comparing the spectral analyses of the extract with those at National Institute for Standard and Technology (NIST, USA) library.
Results
Isolation and screening for bioactive actinobacteria
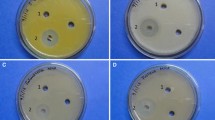
In this study three Streptomyces isolates SUI, SW72IV and SW72VII isolated from Lake Gerio were observed to demonstrate inhibitory activity against test strains such as Bacillus sp., P. aeruginosa ATCC 9077 and S. aureus ATCC 700699.
Cultural, morphological and biochemical characteristics and molecular identification of the actinomycetes
The cultural, morphological and biochemical characteristics of the three selected isolates were carried out on five different International Streptomyces Program (ISP) media (ISP1, ISP2, ISP4, ISP5 and ISP7), starch casein agar and nutrient agar as presented in Table 1.
Abundant growth by Streptomyces sp. SUI was recorded on all the ISP media used as well as on starch casein and nutrient agars. Streptomyces sp. SW72IV had abundant growth on ISP1, ISP5, Starch casein and nutrient agar but poor growth was observed on ISP2, ISP4 and ISP7 media while Streptomyces sp. SW72VII showed abundant growth on ISP1, ISP2, ISP5, ISP7, Starch casein and nutrient agars but poor on ISP4 only. The three isolates were rough in appearance on the media used to study their cultural and morphological characteristics except in some few cases where they appeared smooth and shiny. Additionally, isolate SUI, SW72IV and SW72VII had different diffusible pigments on ISP2, ISP5 and ISP7 media.
Biochemical characteristics of the three isolates as presented in Table 2 showed that isolate SUI metabolized glucose, galactose, fructose, glycerol, arabinose, rhamnose, D-mannitol, meso-inositol, starch and xylose while isolate SW72IV used only five of the sugars that included glucose, glycerol, D-mannitol, meso-inositol and xylose. However, isolate SW72VII did not ferment any of the sugars used. Streptomyces sp. SUI was positive to catalase, urease, gelatinase,oxidase and Gram’s reaction but negative to Voges-Proskauer. However, Streptomyces sp. SW72IV showed reactions to catalase, Voges Proskauer, gelatinase and Gram staining but negative to methyl red, urease and oxidase. Streptomyces sp. SW72VII was only positive to catalase, gelatinase and Gram reaction but negative to methyl red, Voges Proskauer, urease and oxidase.
The 16S rRNA gene nucleotide sequences were used to identify the three actinobacteria isolates. Isolates SUI, SW72IV and SW72VII showed similarity indices of 97%, 99% and 99% making them to be closely related to S. acrimycini strain NBRC 12736, S. pseudogriseolus strain NRRL B-3288 and S. fradiae strain NBRC 12773 respectively. The three isolates have resemblances to other strains of Streptomyces strains at Genbank (Fig. 1). The 16S rRNA gene sequences of isolate SUI (Accession no MT584797), isolate SW72IV (Accession no MT584818) and isolate SW 7VII (Accession no MT584816) were deposited at NCBI GenBank. The results of the gel electrophoresis of the 16S rRNA gene were positive for the three strains of Streptomyces under study. The bands were estimated to be between 1.3 and 1.4 kb in size as shown in Fig. 2.
Antimicrobial activity of Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72sVII
The antimicrobial activity of the three strains presented in Table 3 showed that all the strains possessed antimicrobial activity against both Gram positive and Gram negative bacteria, C. albicans and A flavus making them to be broad spectrum in nature. The data presented in Table 3 indicated that Streptomyces sp. SW72VII was more active against the test strains with zones of inhibition of 20.4 mm against Bacillus sp. and 46.3 mm against P. aeruginosa ATCC 9077. These were quite higher than 22.3–25.1 mm recorded with Gentamicin, the positive control. The antifungal activity was highest against C. albicans and A. flavus with extract of Streptomyces sp. SW72IV with zones of inhibition of 42.2 mm and 29.4 mm respectively while the lowest zone of inhibition were 20.3 mm and 24.2 mm. The negative control, ethyl acetate had no effect on the test strains.
The UV–Vis, IR and GC–MS spectra
The UV–Vis spectra of the bioactive compounds synthesized by the three strains of actinobacteria showed the highest peaks of UV–Vis absorption between 241 and 251 nm (Fig. 3). However, a smaller noticeable absorption was observed at wavelength 287 nm for Streptomyces sp. SW72IV.
IR spectra of the compounds present in the partially purified ethyl acetate extracts of the three strains showed absorption bands of wave numbers that ranged between 604 and 633 cm−1. These wave numbers indicated the presence of aliphatic bromo compounds, 760–999 cm−1 showed the presence of aromatic ring and plane compounds and peaks that ranged between 3412 and 3685 cm−1 pointed to the presence of alcohol and hydroxyl functional groups. Similarly, wave bands that were between 1714 and 1722 cm−1 were observed in the spectra of the ethyl acetate extracts of the three actinobacteria strains studied (Fig. 4).
The GC–MS spectra of the partially purified extract characterized and identified thirteen, thirty four and thirty two compounds in extracts of Streptomyces sp. SUI, Streptomyces sp. SW72 IV and Streptomyces sp. SW72 VII respectively (Additional file 1: Tables S1-S3). The abundance of each compounds showed that Elaidic acid, isopropyl ester, C21H42O2 was mostly produced by Streptomyces sp. SUI. Its percentage abundance was 32% while 11-Hexadecenoic acid-15-methyl ester with 0.93%. Furthermore, Octadec-9-noic acid, C18H34O2 was abundantly synthesized by Streptomyces sp. S72IV with 17.44% while 9-cycloheptadecen-1-one, (Z), C17H30 and 3-Eicosene, (E) both with 0.02% were least produced by the strain. Streptomyces sp. S72VII produced more of 2, 3-dihydroxy propyl elaidate C21H40O4. It had 10.0% abundance and the least synthesized was Cyclopentacontane acetic acid, C7H12O2. These three major compounds have been reported to be biologically active exerting different functions as presented in Additional file 1: Tables S1-S3. The chemical characteristics as well as the biological properties of most of the identified compounds are presented in (Additional file 1: Tables S1-S3) for Streptomyces sp. SUI, Streptomyces sp. SW72 IV and Streptomyces sp. SW72 VII respectively.
Discussion
Isolation and screening for bioactive actinobacteria
Several studies carried out on Lake Gerio are majorly on these two, fishing and dry season farming (Ekundayo et al. 2014; Sogbesan et al. 2018). However, no report has been presented in exploring the actinobacteria present and their bioactive compounds that could be of benefits to man. Different environment especially marine and desert have been reported to harbor actinomycetes that are sources of potent bioactive compounds (Valli et al. 2012; Nithya et al. 2018). The same could be said of Lakes which has been explored for its bioactive compounds from Actinomycetes (Gebreyohannes et al. 2013).The run off from surrounding farm areas during raining season could be a source of organic wastes which might have supported the proliferation of bacteria in the Lake. This was earlier mentioned by Achife et al. (2021). In this study, three strains of Streptomyces were observed to be active against both Gram positive and negative bacteria, which confer them of pharmaceutical importance. Gebreyohannes et al. (2013) had previously mentioned the antimicrobial activity of actinobacteria isolated from Lake Tana in Ethiopia against both Gram positive and Gram negative bacteria. Lakes being reservoir of actinobacteria has been mentioned earlier by Gebreyohannes et al. (2013); Benhadj et al. (2019). Our findings were similar to the earlier reports by these authors who reported antibacterial activity of the Streptomyces species they worked with. Earlier submissions from Cho et al. (2012) and Sudha et al. (2018) showed that antibacterial activities of Streptomyces species isolated from the environment were reported against pathogens such as P. aeruginosa, B. cereus and Klebsiella pnuemoniae as well as methicillin and vancomycin resistant S. aureus.
Cultural, morphological and biochemical characteristics and molecular identification of the actinomycetes
Abundant growth of Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII were observed in almost all the ISP and casein agar and nutrient agar used for cultural study. These observations were slightly different from the report of Singh et al. (2018) who reported moderate and slight growth on the same ISP media for a strain of Streptomyces. A similar pattern of growth of abundance and good was mentioned by Daigham and Mahfouz (2021) with the species of Streptomyces they studied. The observations being reported along with previous submissions could be due to ability of the strains to use the media for growth while some of those media with poor or moderate growth might not support the strains for growth. Different aerial and reverse side colors were observed in the three strains studied. Varying colors of the aerial and substrate hyphae as well as melanoid pigments such as pink, red, black, yellowing green have been mentioned by different authors have been reported by Gebreyohaness et al. (2013), Singh et al. (2018), Daigham and Mahfouz (2021). Different aerial and substrate mycelial such as lovry, cream, ivory and metallic brass were observed in this study. The different aerial, substrate and melanoid colors noticed could be attributed to the physiological status of the strains used for the research.
In this study, it was observed that Streptomyces sp. SUI metabolized all carbon and energy sources used for study, Streptomyces sp. SW72IV utilized only seven out thirteen carbon and energy sources while Streptomyces sp. SW72VII did not metabolize any of the sugars as carbon and energy sources. These observations had been reported earlier (Salim et al. 2017; Ganesan et al. 2017; Aharonowitz and Demain 1978). The use of other media nutrients such as organic acids for energy and carbon requirement by actinomycetes was reported by Aharonowitz and Demain (1978) which could be responsible for the non-utilization of any of the sugars used in this study by Streptomyces sp. SWVII. The three strains studied exhibited different enzymatic properties such as catalase, urease oxidase and gelatinase activities. Varying enzyme activities are common to the actinobacteria. Enzymes are essential to the metabolic and physiological activities of these three strains, such as breaking down of macromolecules for growth and development. Sugars utilization and biochemical characteristics could be attributed to genes and physiological conditions of these isolates. The three strains of Streptomyces species SUI, SW72 IV and SW72 VII were filamentous in morphology and positive to Gram’s reaction. These observations were similar to what was previously reported by Ganesan et al. (2017).
A distinct single ampilicon band of 1.3 kb and 1.4 kb of 16S rRNA gene was observed in the three strains studied. The 16S rRNA gene is the most studied gene used in the identification of bacteria up to species level. The sizes of the bands recorded in this study were within the range of 1.2 kb and 1.5 kb reported by Kumari et al. (2019) and Escalante-Rendiz et al. (2019). The slight difference noticed could be due to the level of purity of the extracted DNA.
Antimicrobial activity of the three Streptomyces species
The three actinobacteria strains being reported exhibited antibacterial activity against both Gram positive and Gram negative bacteria, C. albicans and A. flavus making them to be broad spectrum in nature. However, Gram positive bacteria test strains were more sensitive to the inhibitory activity of the three Streptomyces studied. This could be attributed to the differences in the cell wall constituents. Gram negative test bacteria are less sensitive due to the presence of outer lipopolysaccharide layer, a hydrophobic material which renders the cell wall impermeable for lipophilic substances (Shirling and Gottlieb 1966a, b; Kim et al. 1994). Also the observed difference in the susceptibility of the Gram positive and Gram negative bacteria test organisms could due to secretions of different metabolites by the strains as an entity endowing them with broad spectrum activity (Benhadj et al. 2019). The findings in this work were similar to the previous reports of Ganesan et al. (2017) and Gebreyohaness et al. (2013) that stated the antimicrobial activity of isolated actinobacteria strains against different bacterial test strains. The synergistic antimicrobial activity of the bioactive compounds synthesized by the Streptomyces sp. SW72VII could be responsible for the higher inhibitory activity compared to gentamicin. Furthermore, the extract produced by the three strains exhibited inhibitory activity against C. albicans and A. flavus. Our findings are similar to previous report of Hadizadeh et al. (2015) who stated the inhibition of A. fumigatus by extract of S. rochei HF391. Sarika et al. (2021) reported the isolation of Streptomyces felleus BHPL-KSKU5 with anticandidal property. Additionally, the reported bioactivities of secondary metabolites synthesized by the three strains of Streptomyces used in this study showed that these strains possessed antifungal, antioxidant, anticancer, anti-inflammatory properties.
The UV–Vis, IR and GC–MS spectra
Though the UV absorption recorded ranged between 241 and 251 nm wavelengths, the maximum absorption was recorded at 251 nm for the partially purified ethyl acetate extract of the Streptomyces SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII which suggested the presence of chromone-like nucleus structure in the bioactive compounds produced by the three strains. This was close to the earlier report of Singh et al. (2012) on the metabolites obtained from S. levis RS25. The strains was said to inhibit bacteria. Also, a smaller but noticeable absorption at wavelength 287 nm in the partially purified ethyl acetate extract of Streptomyces sp. SW72 VI might indicate the presence of another major compound. This absorption value was similar to that of extract from the strain G614C1 reported by Maleki and Maschinchian (2011) as well as to the report on Streptomyces isolated from soil samples of South-Eastern Serbia by Ilić et al. (2005) which showed maximum absorbance peaks of UV–Vis spectral data ranged between 215 and 270 nm. The maximum UV peaks observed with the compounds present in the partially purified extracts indicate high polyene nature of the bioactive compounds synthesized by the strains. Different UV–Vis maximum absorption has been absorbed at wavelength such as 213.5 nm, 225 nm, 263.5 nm, 324 nm and 529 nm (Ignacimuthu et al. 2017) while Charousova et al. (2018) observed maximum absorption at wavelengths 268 nm and 364 nm. According to Sudha et al. (2018) several metabolites of microbial origin exhibit characteristics absorption spectra in the UV–Vis region which can be give an insight into the identification of the compounds when the spectra are compared with the spectra documented in the library.
The bands recorded based on their wavelength showed that carbonyl groups functional groups (ketone and aldehyde), esters and carboxylic acids were present in the extracted compounds (Coates 2000; Singh et al. 2018). Functional groups such as aliphatic, silicon oxy, carboxylic acid, hydroxyl and carbonyl were identified in the extract (Ignacimuthu et al. 2018; Adesalu et al. 2016; Cetinyaka et al. 2020). The observed similarities could be attributed to similar bioactive compounds synthesized by these strains of Streptomyces.
Fermentation conditions such as carbon, nitrogen sources, temperature and pH to a large extent influence the production of metabolites by different strains of actinomycetes. A wide range of secondary metabolites that possessed different biological activities were present in the ethyl acetate extract of the three strains of Streptomyces studied in this work. The report being presented in this study was in agreement with the report of Elbendary et al. (2018) who also mentioned that bioactivity of actinobacteria was due to the presence of different bioactive molecules.
Conclusion
The three strains of Streptomyces isolated from Lake Gerio, in Yola, Adamawa State, Nigeria Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII exhibited broad spectrum antibacterial activity. The UV–Vis and IR spectra showed that the bioactive compounds produced by the three strains were unsaturated and polyene in nature. Elaidic acid isopropyl ester, Octadec-9-enoic acid and 2, 3-dihydoxypropyl elaidate was produced in abundance by Streptomyces sp. SUI, Streptomyces sp.SW72IV and Streptomyces sp. SW72VII respectively. These compounds have been reported to possess biological properties such as antimicrobial, antihypertensive, antioxidant and anticancer. The three strains Streptomyces sp. SUI, Streptomyces sp.SW72IV and Streptomyces sp. SW72VII can be used in the production of pharmaceuticals, particularly antibacterial and other bioactive compounds needed in cosmetics and food industries.
Availability of data and materials
The data that support the findings of this study are available at NCBI GenBank database. The 16S rRNA nucleotide sequences of the three strains used in this study can be accessed through the accession number MT584797 for Streptomyces sp. SUI, MT584818 for Streptomyces sp. SW72IV and MT584816 for Streptomyces sp. SW72VII.
Abbreviations
- NCBI:
-
National center for biotechnology information
- UV–Vis:
-
Ultra violet–visible
- IR:
-
Infrared
- GC–MS:
-
Gas chromatography-mass spectrophotometer
- ATCC:
-
American type culture collection
- RNA:
-
Ribonucleic acid
- DNA:
-
Deoxyribonucleic acid
- CDC:
-
Center for diseases and control
- g:
-
Gram
- mL:
-
Milliliter
- μL:
-
Microliter
- mg:
-
Milligram
- h:
-
Hour
- nm:
-
Nanometer
- cm−1 :
-
Reciprocal centimeter
- Kb:
-
Kilobase
- PCR:
-
Polymerase chain reaction
- MEGA:
-
Molecular evolutionary genetics analysis
- dNTP:
-
Deoxyribose nucleoside triphosphate
- ISP:
-
International Streptomyces program
References
Abdelhamid MS, Kondratenko EL, Lomteva NA (2015) GC-MS analysis of phytocomponents in ethanolic extract of Nelumbo nucifera seeds from Russia. J Appl Pharm Sci 5:115–118
Abd-Ellatif AYS, Abdel-razek AS, Hamed A et al (2019) Bioactive compounds from marine Streptomyces sp.: Structure, identification and biological activities. Vietnam J Chem. 57:628–635
Abiodun JA, Miller JW (2007) Assessment of Gerio Lake fishery for enhanced management and improved fish production. J Appl Sci Manag 11:11–14
Achife CE, Ijah UJJ, Bala JD, Oyeleke SB (2021) Microbial population of soil and water around petroleum Depot Suleja, Nigeria and their hydrocarbon utilization. Int J Life Sci Biotech 4:90–113
Adesalu TA, Tenemu TO, Julius ML (2016) Molecular characterization, lipid analysis and GC-MS determination of bioactive compounds identified in a West African strain of the green alga Oedogonium (Chlorophyta). J Pharm Phytochem 5:01–06
Adeyemi MA, Ekunseitan DA, Abiola SS, Dipeolu MA, Egbeyale LT, Sogunle OM (2017) Phytochemical analysis and GC-MS determination of Lagenaria breviflora R fruit. Int J Pharm Phytochem Res 9:1045–1050
Aharonowitz Y, Demain AL (1978) Carbon catabolite regulation of cephalosporin production in Streptomyces clavuligerus. Antimicro Agents Chemothe 14:150–164
Ahmed M, Peiwen Q, Gu Z et al (2020) Insecticidal activity and biochemical composition of Citruluss colocynthis, Cannabis indica and Artemisia argyi extract against cabbage aphid (Brevicoryne brassicae L.). Sci Rep 10:552
Ajayi GO, Olagunju OA, Martins OC (2011) Gas chromatography-mass spectrometry analysis and phytochemical screening of ethanolic root extract of Plumbago zeylanica Lin. J Med Plant Res 5:1756–1761
Ansari A, Ali A, Asif M (2017) Biologically active pyrazole derivatives. New J Chem 41:16–41
Anthony BO, Oladipo AL, Adeola AS, Folorunsho OS, Azeez II, Andy RO (2013) Volatile constituents antioxidant and insecticidal activities of essential oil from leaves of Thaumatococcus danielli (benn) Benth. From Nigeria IOSR J Pharm 3:1–5
Arora G, Sharma S, Sahni T, Sharma P (2018) Antioxidant and antimicrobial activity of 2-phenyl-1-H-indoles and Benzimidazoles. Indian J Pharm Sci 80:739–744
Atta HM, Ahmad MS (2009) Antimycin-A antibiotic Biosynthesis produced by Streptomyces sp. AZ-AR-262: Taxonomy, fermentation, purification and biological activities. Aust J Basic Appl Sci 3:126–135
Aukema H, Campbell L (2011) Oil nutrition and utilization chemistry. Production, processing and utilization. Elsevier, Amsterda, pp 245–280
Barathikannan K, Venkatadri B, Khusro A et al (2016) Chemical analysis of Punica granatum fruit peel and its in-vitro and in-vivo biological properties. BMC Complement Alternative Med 6:264
Barka EA, Vatsa P, Sanchez L, Gavau-Vailant N, Jacquard C, Klenk HP (2016) Taxonomy physiology natural products of actinobacteria. Microbiol Mol Bio Rev 80:1–43
Bauer AW, Roberts CE Jr, Kirby WM (1959) Single disc versus multiple disc and plate dilution techniques for antibiotics sensitivity testing. Antibiot Annu 7:574–580
Benhadj M, Gracemi-Kirane D, Manasria IT, Guebla K, Ahmane Z (2019) Screening of rare actinomycetes isolated from natural wetland ecosystem (Fetzara lake, northeastern Algeria) for hydrolytic and antimicrobial activities. J King Saud Uni-Sci 31:706–712
Berdy J (2005) Bioactive microbial metabolites. J Antibiot 58:1–6
Brintha S, Rajesh S, Penuka R, Santhanakrishnan VP, Gnanam R (2017) Phytochemical analysis and bioactivity prediction of compounds in methanolic extracts of Curculigo orchioides gaestn. J Pharm Phytochem 6:192–197
Center for Disease and Control (CDC) (2019) Antibiotic resistance threats in the United States, US Department of Health and Human Services, CDC, Atlanta, GA, p 2019
Cetinyaka S, Yenidunya AF, Arslan K, Arslan D, Dogan O, Dastan T (2020) Secondary metabolites of Streptomyces griseorubens isolate are predominantly pyrrole and Linoleic acid like compounds. J Oleo Sci 69:1273–1280
Charousova I, Medo J, Hleba L, Javorekova S (2018) Streptomyces globosus DK15 and Streptomyces edreresis ST13 as new producers of factumycin and tetrangomycin antibiotics. Braz J Microbiol 49:816–822
Cho SS, Choi YH, Simkhada JR, Mander P, Park DJ, Yoo JC (2012) A newly isolated Streptomyces sp. CS392 producing three antimicrobial compounds. Bioprocess Biosyst Eng 35:247–254
Chowdury K, Sharma A, Kumar S, Gunjan GK, Nag A, Mandal CC (2017) Colocynth extracts prevent epithelial to mesenchymal transition and stemness of breast cancer cells. Front Pharmacol 8:593–606
Coates J (2000) Interpretation of infrared spectra, a practical of green chemistry into future approach. In: Meyers RA (ed) Encyclopedia of analytical chemistry. John Wiley and Sons Ltd, Chochester, pp 10815–10837
Collins CH, Lyne PM, Grange JM (1995) Amylase production of Streptomyces rimosus TM-55 and their 2-deoxyglucose Mutants. Chin J Microbiol Immunol 28:109–116
Daigham GE, Mahfouz AY (2021) Isolation, characterization and screening of actinomycetes producing bioactive compounds from Egyptian soil. Egypt Pharm J 19:381–390
Dalli AK, Saha G, Chakraborty U (2007) Characterization of Antimicrobial compounds from a common fern, Pterin Biaurita. Indian J Exp Biol 45:285–290
Darbro JM, Miller JG, Mcelfresh JS, Mullens BA (2005) Survey of muscalure [(Z)-9-tricosene] on house flies (Diptera: Muscidae) from field populations in California. Environ Entomol 34:1418–1425
Debananda SN, Sanasam S, Nimaichand S (2009) Screening of actinomycetes isolates from niche habitats in Manipur for antibacterial activity. Am J Biochem Biotechnol 5:221–225
Desbois AA, Smith VJ (2010) Antibacterial free fatty acids: Activities, mechanisms of action and biotechnology potential. Appl Microbiol Biotechnol 85:1629–1642
Dilika F, Bremner PD, Meyer JJ (2000) Antibacterial activity of linoleic and oleic acids isolated from Helichrysum peduncalatum: a plant used during circumcision rites. Fitoterapia 71:450–452
Duke J (1998) Duke’s phytochemical and ethnobotanical database. www.ars.grin.gov/duke/. Accessed 30 Aug 2020.
Ekundayo MT, Sogbesan OA, Haruna AB (2014) Study of fish exploitation pattern of Lake Gerio, Yola, Adamawa State. Nigeria J Surv Fish Sci 1:9–20
Elbendary AA et al (2018) Isolation of antimicrobial producing actinobacteria from soil samples. Saudi J Biol Sci 25:44–46
Escalante-Rendic O, de-la-Rosa-Garcis S, Tapia-Tussell R, Martin J, Reyes F, Vicente F et al (2019) Molecular identification of selected Streptomyces strains isolated from Mexican tropical soils and their anti-candidal activity. Int J Environ Res Pub Health 16:1913
Fawole MO, Oso BA (2004) Characterization of bacteria: laboratory manual of microbiology, 4th edn. Spectrum Book Ltd, Ibadan, pp 24–33
Fustero S, Sánchez-Roselló M, Barrio P, Simón-Fuentes A (2011) From 2000 to Mid- 2010: a fruitful decade for the synthesis of pyrazoles. Chem Rev 111:6984–7034
Ganesan P, Reegan AD, Rajendra HAD et al (2017) Antimicrobial activity of some actinomycetes from Western Ghats of Tamil Nadu. India Alex J Med 53:101–110
Gary JB, Richard GV (2003) Insect pheromone biochemistry and molecular biology: The biosynthesis and detection of pheromones and plant volatiles. Academic Press, Cambridge, p 232
Gebreyohannes G, Moges F, Sahile S, Raja N (2013) Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac J Trop Biomed 3:426–435
Goodfellow M, Williams ST (1983) Ecology of actinomycetes. Ann Rev Microbiol 37:189–216
Guzman JD (2014) Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 19:19292–19349
Hadizadeh S, Forootanfar H, Shahidi BGH et al (2015) Antifungal activity of terrestrial Streptomyces rochei strain HF391 against clinical-azole resistant Aspergillus fumigatus. Curr Med Myc 1:19–24
Hahn JU, Assenmacher-Maiworm HM, Koch KH, Pannwitz R, Hebiseh TH, Brock AH (2016) The MAK collect. Occup Health Saf 2016(1):2734–2748
Handhini US, Sudha S, Anusha JV, Manisha S (2018) Isolation, identification and extraction of antimicrobial compounds produced by Streptomyces species from terrestrial soil. Biocat Agric Biotechnol 5:317–321
Harrigan WF, McCance ME (1976) Laboratory methods in microbiology. In: Gibbs BM, Skinner FA (eds) Laboratory methods in microbiology. Academic Press, London, pp 14–23
Hase GJ, Deshmukh KK, Pokharkar RD, Gaje TR, Phananagre ND (2017) Phytochemical studies on Nerium oleander L. using GC-MS. Int J Pharm Phytochem Res. 9:885–891
Hatami S, Sani AM, Yavarmanesh M (2016) Chemical composition and antibacterial activity of organic extra virgin olive oil from Iran. Nutri Food Sci 46:388–395
Hazarika S, Goswami N, Dutta NN, Hazarika A (2002) Ethyl oleate synthesis by porcine pancreatic lipase in organic solvent. J Chem Eng 85:61–68
Hoda S, Gupta L, Shankari J, Gupta AK, Vijayaraghavan P (2020) Cis-9-Hexadecenal, a natural compound targeting cell wall organization, critical growth factor and virulence of Aspergillus fumigatus. ACS Omega 5:10077–10088
Ignacimuthu S, Joseph DB, Gabriel PM, Agastian STP, Aldhadi NA (2017) Identification of antimicrobial compounds from Streptomyces sp. isolated from Western Ghats soil in Tamil Nadu. EC Microbiol. 8:222–231
Ilić SB, Konstantinović SS, Todorović ZB (2005) UV/Vis analysis and antimicrobial activity of Streptomyces isolates. FU Med Biol 12:44–46
Kim C, Lee K, Kwon O, Yoo I, Shimazu A (1994) Selective isolation of actinomycetes by physical treatment of soil sample. K J Appl Microbiol Biotechnol 22:222–225
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549
Kumari S, Sharma U, Thakur A, Krishna R, Kumar S (2019) 16S rRNA gene sequencing and amplification of gelatinase-producing Streptomyces strains of fruit orchard. IOSR J Biotechnol Biochem (IOSR-JBB) 5:25–31
Küster E, Williams S (1964) Media for the isolation of streptomycetes: starch casein medium. Nature 202:928–929
Lotfy MM, Hassan HM, Hetta MH, El-Gendy AO, Mohammed R (2018) Di-(2-ethylhexyl) phthalate, a major bioactive metabolite with antimicrobial and cytotoxic activity isolated from River Nile derived fungus Aspergillus awamori. Beni-Suef Uni J Bas Appl Sci 7:263–269
Makhafola TJ, Elgorashi EE, McGraw LJ, Awouafack MD, Verschaeve L, Eloff JN (2017) Isolation and characterization of the compounds responsible for the antimutagenic activity of Combretum microphyllum (combretaceae) leaf extract. BMC Complement Alternative Med 17:446
Maleki H, Maschinchian O (2011) Characterization of Streptomyces isolates with UV, FTIR spectroscopy and HPLC analyses. BioImpacts 1:47–52
Malibari AA (1991) Isolation and screening of antibiotic producing Streptomycetes from Western region soils of Saudi Arabia. J Kingdom Acad Sci 3:31–42
Mehta S, Atpathy A, Gupta RK (2014) Evaluation of nutritional, phytochemical, antioxidant and antibacterial activity of dried plum (Prunus domestica). J Pharm Phytochem 3:166–171
Monciardini P, Sosio M, Cavaletti L, Chiocchini C, Donaldio S (2002) New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol 42:419–429
Nandhini US, Sangareshwari S, Kumari L (2015) Gas chromatography-mass spectrometry analysis of bioactive constituents from three marine Streptomyces. Asian J Pharm Clin Res 8:244–246
Nithya K, Muthukumar C, Biswas B et al (2018) Desert actinobacteria as a source of bioactive compounds production with a special emphasis on pyridine-2,5-diacetamide, a new pyridine alkaloid produced by Streptomyces sp. DA3–7. Microbiol Res 207:116–133
Omotoyinbo BI, Aje AE, Kolapo OS, Alagbe OV (2018) Bioactive constituents of essential oils from Khaya senegalensis (Desr) bark extracts. Am J Chem Biochem Eng 2:50–54
Oskay M (2009) Antifungal and antibacterial compounds from Streptomyces strains. Afric J Biotechnol 8:3007–3017
Prescott LM, Harley JP, Klein DA (1991) Microbiology, 2nd edn. WMC Brown Publishers, New York, p 912
Qinn GA, Banat AM, Abdelhammed AM, Banqt IM (2020) Streptomyces from traditional medicine sources of new innovations in antibiotic discovery. J Med Microbiol 9:1040–1048
Rahman MM, Ahmad SH, Mohamed MTM, Abrahaman MZ (2014) Antimicrobial compounds from leaf extracts of Jatropha curcas, Psidium guajava, and Andrographis paniculata. Sci World J 2014:635240–240
Ramesh R, Dhanaraji TS (2016) Identification of bioactive compounds in the ethyl acetate extract of Terminalia arjuna root by GC-MS analysis. Int J Recent Sci Res 7:9747–9751
Salim FM, Sharmili SA, Anbumalarmathi J, Umamaheswari K (2017) Isolation, molecular and identification of antibiotic producing actinomycetes from soil samples. J Appl Pharm Sci 7:069–075
Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448
Sanglier JJ, Haang H, Huck TA, Fehr T (1996) Review of actinomycetes compounds 1990–1995. Expert Opin Investig Drugs 5:207–223
Sarika K, Sampath G, Govindarajan RK et al (2021) Antimicrobial and antifungal activity of soil actinomycetes isolated from coal mines sites. Saudi J Biol Sci. 28:3553–3558
Schneider I, Kressel G, Meyer A, Krings U, Berger RG, Hahn A (2011) Lipid lowering effects of Oyster mushroom (Pleurotus ostreatus) in human. J Funct Food 3:17–24
Sebak M, Saafan AE, Abdelghani S, Bakeer MAS, El-Gendy AO (2021) Isolation and optimized production of putative antimicrobial compounds from Egyptian soil isolate Streptomyces sp. MS10. Beni-Suef Uni J Basic Appl Sci 10:8
Sharma P, Thakur D (2020) Antimicrobial biosynthetic potential and diversity of cultrurable soil actinobacteria from forest ecosystems of Northeast India. Sci Rep 10: 4104
Sharma V, Chitranshi N, Agarwal AK (2014) Significance and biological importance of pyrimidine in microbial world. Int J Med Chem 2014:1–31
Shirling EB, Gottlieb D (1966b) Methods, classification, identification and description of Genera and species. The William and Wilkin Company, Baltimore, pp 261–292
Shirling EB, Gottlieb D (1966a) Methods for characterization of Streptomyces. Int J Syst Bacteriol 16:313–340
Siddharth S, Vittal RR, Wink J, Steinert MC (2020) Diversity and bioactive potential of actinobacteria from unexplored regions of Western Ghats. India Microorganisms 8:225
Singh V, Tripathi C, Bihari V (2008) Production and optimization of an antifungal compound from Streptomyces capoamus MTCC 8123. Med Chem Res 17:94–102
Singh N, Rai V, Tripathi C (2012) Production and optimization of oxytetracycline by a new isolate Streptomyces rimosus using response surface methodology. Med Chem Res 21:3140–3145
Singh V, Haque S, Khare S et al (2018) Isolation and purification of antibacterial compound from Streptomyces levis collected from soil sample of north India. PLoS ONE 13:e0200500
Smist M, Kwiecien H, Krawczyk M (2016) Synthesis and antifungal activity of 2H–1,4-benzoxazin-3 (4H)-one derivatives. J Environ Sci Health Part B Pesticides Food Contaminants Agric Wastes 51:393–401
Sogbesan OA, Tukura EE, Ekundayo MT (2018) Parasitic infection and prevalence of Clarias geriepinus in Lake Gerio, Yola, Adamawa State. MOJ Anat Physiol 5:Y376–Y381
Sudha SKS, Usha N, Girija V (2018) Chromatography-based partial purification and characterization of antimicrobial compound isolated from a terrestrial Streptomyces. Asian J Pharm Clin Res 11:2
Tonisi S, Okaiyeto K, Hoppe H, Mabinya LV, Nwodo UU, Okoh AI (2020) Chemical constituents, antioxidant and cytotoxicity properties of Leonotis leonurus used in the folklore management of neurological disorders in the Eastern Cape. South Africa Biotech 10:141
Tripathi C, Praveen V, Singh V, Bihari V (2004) Production of antibacterial and antifungal metabolites by Streptomyces violaceusniger and media optimization studies for maximum metabolites. Med Chem Res 13:790–799
Tumlinson J, Brennan MM, Doolittle RE et al (1989) Identification of a pheromone blend attractive to Manduca sexta (L) males in a wind tunnel. Arch Insect Biochem Physiol 10:255–271
Upadhyay RK, Jaiswal G (2007) Evaluation of biological activities of Piper rugrumi oil against Tribolium castaneum. Bull Insectol 60:57–61
Valli S, Suvathi SS, Aysha OS, Nirmala P, Vinoth KP, Reena A (2012) Antimicrobial potential of Actinomycetes species isolated from marine environment. Asian Pac J Trop Biomed 2:469–473
Wintola OA, Afolayan AJ (2017) Chemical constituents and biological activities of essential oils of Hydnora africana thumb used to treat associated infections and diseases in South Africa. Appl Sci 7:443
Wisneski HH, Yates RL, Davis HM (1982) Gas chromatographic determination of synthetic musk (7-Acetyl-6-ethyl-1,1,4,4-tetra methyl tetralin in fragrances. J Assoc Anal Chem 55:598–601
Wu RY (1984) Studies on the Streptomyces SC4II-Taxonomical and biological characteristics of Streptomyces strain SC4. Bot Bull Acad Sci 25:111–123
Yakubu MB, Lawal AO, Jasper EE (2018) GC-MS Analysis of ethyl acetate fraction of the whole plant extract of Pysochoriste perrottetii (Acanthaceae). Nig J Chem Res 23:52–58
Acknowledgements
The authors wish to appreciate the support received from the Research and Development Unit of Modibbo Adama University of Technology, Yola and TET-fund, Abuja, Nigeria to carry out this work.
Funding
Author OM appreciates the funding of this research by TET-fund, Abuja, Nigeria and the support received from Research and Development Unit of Modibbo Adama University of Technology, Yola, Adamawa State, Nigeria especially the buying of basic chemicals and consumables used for this study.
Author information
Authors and Affiliations
Contributions
Author OM conceived the idea, OM, MI and FV carried out the laboratory work, collection and analysis of data, OM drafted the original manuscript, proofread and corrected by MI and FV. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Supplementary Table S1-S3. Bioactive compounds identified by GC-MS produced by Streptomyces sp. SUI, Streptomyces sp. SW72IV and Streptomyces sp. SW72VII.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adeyemo, O.M., Ja’afaru, M.I. & Adams, F.V. Isolation, characterization, antimicrobial and other bioactivity profiles of three Streptomyces strains isolated from Lake Gerio, Yola, Adamawa State, Nigeria. Bull Natl Res Cent 45, 147 (2021). https://doi.org/10.1186/s42269-021-00606-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-021-00606-x