Abstract
Background
Determining the spatial distribution of tree heights at the regional area scale is significant when performing forest above-ground biomass estimates in forest resource management research. The geometric-optical mutual shadowing (GOMS) model can be used to invert the forest canopy structural parameters at the regional scale. However, this method can obtain only the ratios among the horizontal canopy diameter (CD), tree height, clear height, and vertical CD. In this paper, we used a semi-variance model to calculate the CD using high spatial resolution images and expanded this method to the regional scale. We then combined the CD results with the forest canopy structural parameter inversion results from the GOMS model to calculate tree heights at the regional scale.
Results
The semi-variance model can be used to calculate the CD at the regional scale that closely matches (mainly with in a range from − 1 to 1 m) the CD derived from the canopy height model (CHM) data. The difference between tree heights calculated by the GOMS model and the tree heights derived from the CHM data was small, with a root mean square error (RMSE) of 1.96 for a 500-m area with high fractional vegetation cover (FVC) (i.e., forest area coverage index values greater than 0.8). Both the inaccuracy of the tree height derived from the CHM data and the unmatched spatial resolution of different datasets will influence the accuracy of the inverted tree height. And the error caused by the unmatched spatial resolution is small in dense forest.
Conclusions
The semi-variance model can be used to calculate the CD at the regional scale, together with the canopy structure parameters inverted by the GOMS model, the mean tree height at the regional scale can be obtained. Our study provides a new approach for calculating tree height and provides further directions for the application of the GOMS model.
Similar content being viewed by others
Introduction
Tree height is one of the main forest vertical structural parameters, and it can reflect the overall state of the forest structure. Moreover, tree height is the main input parameter for estimating forest volume and forest above-ground biomass (AGB). It also represents a natural characteristic of the allometric growth mechanism and an indicator of forest total resource utilization, biomass productivity, spatial distribution, death, rebirth, etc. (Enquist et al. 1998; Enquist et al. 2009; Enquist and Niklas 2001; Muller-Landau et al. 2006; West et al. 2009). Research on tree height has far-reaching significance for the study of forest ecosystems.
The main methods of obtaining tree height in forest studies include field measurements, statistical model estimates, and physical model inversions based on field-measured data or remote sensing data. A total station device is an instrument that is often used to measure tree height in the field and it provides direct, current, accurate and reliable data for determining the three-dimensional coordinates of a tree. Although the field measurement method can obtain tree height with high accuracy, the detected area is limited due to the substantial technical requirements and material resource costs. In forest science studies, statistical regression methods have been widely used to investigate vegetation parameters of forests, and according to the principles of tree growth, the tree height in a specific zone is highly correlated with numerous forest parameters, including the diameter at breast height (DBH) (Ercanlı 2020) and stand age (Xiong et al. 2016). Richard (Carmean and Lenthall 1989; Payandeh 1974; Payandeh and Wang 1994a), Logistic (Chen et al. 1998; Nigh and Sit 1996; Thrower and Goudie 1992; Wang and Klinka 1995), and Weibull (Payandeh and Wang 1994b; Yang et al. 1978) are the most frequently used statistical models to estimate tree height. However, these statistical models are primarily based on field measurements, and obtaining the stand age and DBH at the regional scale is unfeasible. Statistical models are not well-suited for calculating tree height at the regional scale. With the development of remote sensing science and technology, remote sensing data have been widely used to retrieve tree height.
Laser radar technology is the main method for obtaining high-resolution tree height data, and researchers have developed numerous algorithms to derive tree height from LiDAR data (Nelson et al. 1997; Nilsson 1996). Airborne laser scanning (ALS) provides 3D structure information as well as the intensity of the reflected light and has become established as an important instrument in forestry applications (Edson 2011). ALS has been successfully used to estimate the canopy height, leaf area index (LAI), biomass and other variables (Dubayah et al. 2010; Lefsky et al. 2005; Lefsky et al. 2007; Ma et al. 2014; Riaño et al. 2004). Data from ALS can provide precise individual tree detection (ITD), and researchers use the spectral (Breidenbach et al. 2010; Heinzel and Koch 2012; Leckie et al. 2003) and intensity information (Huo and Lindberg 2020) for ITD studies. Lefsky et al. (2002) showed that together with the remote sensing of topography, the three-dimensional structure and function of vegetation canopies can be directly measured and forest stand attributes accurately predicted. Means et al. (1999) reported that compared with field-measured tree height, large-footprint, airborne scanning LiDAR can be used to precisely characterize stand structure with R2 equal to 0.95. However, the weak penetration of laser pulses in dense forest coverage makes it difficult to obtain the forest canopy vertical structural parameters using this method, and the high cost and the lack of mapping capacity also limit the application of ALS at regional and global scales (Sun et al. 2006; Swatantran et al. 2011). Thus, developing a method to obtain tree height at regional and global scales is critical for improving forest studies and developing long-term strategies for forest ecosystem protection.
The geometric-optical mutual shadowing (GOMS) model increases the suitability of the geometric-optic model for highly dense canopy forests (Li and Strahler 1992) and is particularly suitable at the regional scale. The GOMS model describes the tree canopy 3-D structure and successfully establishes the relationship between forest structure parameters (e.g., average vegetation coverage, average tree height, crown size) and the canopy bidirectional reflection distribution function, yielding the relationship between canopy structure parameters (e.g., clear height, crown radius, forest canopy distribution) and the canopy reflection characteristics (Li and Strahler 1985). Then, forest canopy structural parameters can be inverted by the GOMS model. However, the GOMS model can obtain only the ratio between different canopy structural parameters, such as b/R and h/b, in which R represents the horizontal radius of an ellipsoidal crown, b represents the vertical half axis of an ellipsoidal crown, and h represents the height at which a crown center is located (Li et al. 2015). To obtain tree height, field-measured data and LiDAR data are required to provide the canopy diameter (CD) or clear height (Fu et al. 2011; Ma et al. 2014). Realistically, in tree height studies, high-accuracy field-measured data and LiDAR data are not always available at the regional scale. Easily and cheaply providing CD or other canopy structure parameters as prior knowledge for the tree height calculation process through the GOMS model is an important and meaningful research direction in tree height studies. Therefore, in this study, we attempted to build a method for calculating the CD or clear height with optical remote sensing data instead of field-measured and LiDAR data; then, tree height at the regional scale can be easily obtained using the GOMS model. We learned from the research of Song et al. (2010), who successfully calculated CD by using high-resolution imagery, and applied the CD calculation method to the regional scale. We chose the Dayekou forest study site as a study area, used a semi-variance model to calculate the CD, and then extended this method to the regional scale. Then, combined with the canopy structural parameters inverted by the GOMS model, including b/R and h/b, tree height (H = h + b) could be calculated. The accuracy of the estimated tree height was validated using canopy height model (CHM) data derived from LiDAR data.
Materials and methods
Study site
The study site, Dayekou forest (100° E, 38.6° N), is located in the Qilian Mountain area of Gansu Province, China. The Heihe Basin is the second largest inland river basin of the arid region in northwestern China, with annual precipitation of 140 mm (Li and Xu 2011) in the middle valley. Dayekou is located in the middle valley of the Heihe River Basin, and most of the area is covered by forest and upland meadow. The main vegetation types in the Dayekou forest are Picea crassifolia, shrubland and upland meadow, and the dominant forest type is P. crassifolia.
The locations of the field measurement sample plots are shown in Fig. 1. One super sample plot sized 100 m × 100 m is located within the yellow line surrounding the pixels as indicated within the Dayekou site. The super sample plot was divided into 16 parts, each sized 25 m × 25 m. In each small sample plot (B 1–16), all parameters related to trees were measured, including LAI and canopy structure parameters (tree height, canopy diameter, etc.). The field-measured canopy structure parameters measured in these super sample plots are described in the section of field-measured data. The super sample plots were relatively homogeneous.
Standard false color image (SPOT-5) of the experimental sites. The super sample plot is outlined in black. The area outlined in yellow of this map is the same as that of the CCD image shown in Fig. 2
Data foundation
Field-measured data
Field measurements in the super sample plots were performed in June 2008. The measured geometrical structural parameters included the horizontal radius of the tree crown (R), tree height (H), clear bole height (h), and DBH. The height of each tree in the super sample plots was measured via a laser altimeter (TruPulse 200, Laser Technology Inc. (LTI), Norristown, PA USA). Field-measured data (Chen and Guo 2008) were used to construct a prior knowledge database of the canopy structural parameters for the GOMS model.
The protocols for each instrument used in the sample plots and the sample-plot layouts were as described in a previous study (Fu et al. 2011).
Bidirectional reflectance data and high spatial remote sensing data
In this research, both bidirectional reflectance data and optical high spatial resolution remote sensing data were used. The detailed information on the datasets is provided in Table 1. Moderate-resolution Imaging Spectro Radiometer (MODIS) and Multi-angle Imaging Spectro radiometer (MISR) reflectance products were used to build the multi-angle bidirectional reflectance (BRF) datasets (Fu et al. 2011; Li et al. 2015), which were the input data in the canopy structure parameter inversion process performed by the GOMS model. SPOT-5 data can be used to acquire the spectral information (G, C, Z and T) (Fu et al. 2011), and we also used the SPOT-5 image to perform the supervised classification with the Environment for Visualizing Images (ENVI; Exelis, Inc., Boulder, CO, USA) to provide the landcover information for the CD calculation process in the section of tree height and CD results derived from the CHM data (Fig. 11). Airborne CCD multi-band imagery (Li et al. 2017; Xiao 2018) was used to calculate the spatial variation in the study area with the semi-variance model in the section of tree height and CD results derived from the CHM data (Fig. 2) (Song et al. 2010). Airborne LiDAR data provided the CHM information to estimate the accuracy of the tree height inversion results.
Methods
GOMS model and inversion strategy
The GOMS model was constructed based on the Li-Strahler geometric-optical model (Li and Strahler 1992), which assumes that the reflectance of a pixel can be modeled as a sum of the reflectance of its individual scene components weighted by their respective areas within the pixel (Li and Strahler 1985) and that the vegetation canopy bidirectional reflectance distribution function (BRDF) characteristics at the pixel scale can be explained by the geometric-optical principle. The sensors receive the ground reflection and the crown reflection in the field of view A (“A” is the assumption that the area of the field of view is A).
Considering the 3-D forest canopy structural parameters, the influence of sky light, and multiple scattering, the received signal of A can be defined as a combination of the four area-weighted components:
where S refers to bidirectional reflectance factor (BRF); Kg, Kc, Kz, and Kt are the proportions of sunlit background, sunlit crown, shaded background, and shaded crown, respectively; and G, C, Z and T are the contributions of the sunlit background, sunlit crown, shaded background, and shaded crown, respectively (Li and Strahler 1986).
Assuming that the tree crown shape is ellipsoidal (Fig. 3a), Kg, Kc, Kz and Kt can be expressed by a combination of the forest canopy structural parameters such as R, b, h and n (the number of crowns per unit area).
a Forest canopy shape as an ellipsoid. b A single sphere viewed at position v and illuminated at position i (Li and Strahler 1992). θi and θv are the revised solar zenith angle and view zenith angle, respectively. ∅i and ∅v are the solar azimuth and view azimuth, respectively. ∅i − ∅v is the azimuthal difference between the illumination and viewing directions. τi and τv are the sunlit shadow and viewed shadow, respectively. The shaded area is the mutual shadowing area of the sunlit shadow and viewed shadow
In the GOMS model, the ellipsoid model is simplified into a spheres model (Fig. 3b); then, Kg, Kc, Kz and Kt can be expressed as:
where
and O(θi, θv, ∅i − ∅v) is the shaded area in Fig. 3b. ∅i and ∅v are the solar azimuth and view azimuth, respectively, and θi and θv are the revised solar zenith angle and view zenith angle, respectively:
where θi′ and θv′ are solar zenith angle and view zenith angle, respectively.
Then, the GOMS model can be expressed by the function below:
where nR2 represents the crown coverage condition per unit area in the nadir observation, b/R affects the crown coverage density in the non-nadir direction; h/b affects the outward width of the hot spot; and ∆h/b describes the discrete degree of the crown height distribution and affects the bowl-shape of the BRDF (∆h is the variance of the h distribution in one pixel) (Li et al. 2015). θs and ∅s are the local slope and aspect, respectively. θi, ∅i, θv and ∅v are the solar zenith angle, solar azimuth, view zenith angle, and view azimuth, respectively (Fu et al. 2011; Ma et al. 2014). In this study, we assume that the reflected intensities of the shadow on the ground and on the canopy are the same (i.e., Z equals T). Thus, the model is simplified with three area-weighting components (G, C and Z).
The multi-stage, sample-direction dependent, target-decisions (MSDT) inversion method (Li et al. 1997) was adopted to segment invert the observation data and the parameters in the GOMS model. In this method, the most sensitive observation data were used to invert the most sensitive parameters; then, the previous inversion results were used as the prior knowledge in the next parameter inversion stage. The parameter inversion order is based on the uncertainty and sensitivity matrix (USM), which presents the sensitivity of the parameters to the observational data in different viewing directions. The USM function can be expressed as
where ∆BRF(p, q) is the maximum difference of BRF calculated by the model when only parameter q changes in its uncertainty and other parameters remain fixed, and BRFexp(p) is the BRF calculated by the model at the pth geometry of illumination and viewed with all parameters at their expected values. Based on our previous study (Fu et al. 2011), the inversion order of all the parameters in the GOMS model is RC- > RG- > RZ and NIRC- > (b/R, NIRZ, ∆h/b)- > NIRG- nR2. RC-RG-RZ refers to the BRF information of sunlit crown, sunlit background, and shaded area in the red band, and the NIRC-NIRG-NIRZ refers to the BRF information of the sunlit crown, sunlit background, and shaded area in the near-infrared (NIR) band. Then, the parameters in both the NIR and red bands were used to calculate h/b. From the inversion order results, R (R = CD/2) was not a very sensitive parameter in the GOMS model; thus, using the CD provided by other data sources as prior knowledge in the GOMS model inversion procedure to calculate tree height would not cause substantial error.
Semi-variance model
The semi-variance model is a tool to build the relationship between the underlying scene and the image spatial properties and the image spatial properties can be measured by calculating the spatial variation of a spatial random variable. In a remote sensing image, each digital number (DN) is linked to a unique location on the ground and can be considered the realization of a spatial random function: DNi = f(xi), where DNi is the digital number for the ith pixel, xi is the geographic location vector for the ith pixel, and f is the random spatial function. The DNs of a remotely sensed image can be treated as a spatial random variable. Therefore, the image spatial properties can be estimated by calculating the spatial variation in DN.
A semivariogram (Fig. 4) is a plot of semi-variance against the lag that separates the points used to estimate the semi-variance and can be used to study the spatial properties of the underlying scene (Song 2007).
Typical shape of a semivariogram over a stationary scene (Song 2007)
A semivariogram contains three parameters: the sill, the range and the nugget effect. The sill is the maximum value of semi-variance that presents the total variance of the scene, and it can be calculated by the semi-variance model. The range is the distance at which the semi-variance reaches the sill value, which reflects the scale characteristics of the scene. When the distance between points in space is equal to or greater than the range, these points can be considered to be independent of each other. The nugget effect is the semi-variance at lag zero.
The semi-variance model is defined as follows:
where γf(h) is the semi-variance for points with lag h in space, f(x) is the realization of a spatial random function at location x, f(x + h) is the realization of the same function at another point with lag h from x, and E(.) denotes the mathematical expectation (Song et al. 2010).
Based on the semi-variance model and the theory of Jupp et al. (1988, 1989), the disc scene model was developed, which simplifies the representation of a forest scene. The model assumes a scene that is composed of discs, and the brightness value of a disc does not change in overlapped areas. The model is constructed from the relationship between the scene structure and the spatial characteristics of image DNs. Based on the disc scene model, Song et al. (Song 2007; Song et al. 2002; Song and Woodcock 2003) developed a model that relates the ratio of the sill at two spatial resolutions to the diameter of the object as follows:
where Dp1 and Dp2 are the pixel sizes of the two spatial resolutions; D0 is the diameter of the object (forest CD); and Cz1 and Cz2 are the sills of the regularized semivariograms at spatial resolutions Dp1 and Dp2, respectively. γz1z2 is used to denote the ratio (Cz1/Cz2) described in the latter part of the paper (e.g., γ12 denotes the ratio of the image semi-variance at a spatial resolution of 1 m to that at 2 m).
‘A’ represents the object area:
T(t) represents the overlap function for the objects in the scene:
where
In Eq. (13), the ratio of the sill of the regularized variogram of two different spatial resolutions would be solely determined by the scene structure, which is independent of the brightness value of the pixels. Therefore, the ratio of image variances can be used to estimate the tree crown size across sensors and sites.
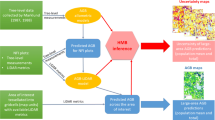
Flowchart of the methods
Figure 5 shows a flowchart of our method, which consists mainly of three parts: the first for the CD calculation process based on the semi-variance model, the second for the tree height estimation process using the CD results from part 1 along with the inversion results obtained from the GOMS model, and the third for the tree height accuracy validation process.
In the CD calculation process, we applied the CD estimation process of Song et al. (Song 2007; Song et al. 2002; Song and Woodcock 2003) to the Dayekou forest site using the regularized semi-variance model and high spatial resolution CCD imagery. The optimal fitting function between the sill and the field-measured CD was constructed based on the 16 super sample plots. We first cut the 16 sample plots out of the CCD image employing binarization, then resampled the binary results to different spatial resolutions (1, 2, … 6 m), and finally calculated the sill ratio value of the 16 images at a different spatial resolution. Second, we built the function between the field-measured CD and the sill ratio value and selected the best fitting relationship as the optimal fitting function. Using the supervised classification results for the SPOT-5 image, the method was applied first to the experimental small plot and then to the whole image. We also used the CD derived from the CHM data to analyze the accuracy of the CD data calculated based on the CCD image.
Canopy structural parameters could be inverted by the GOMS model, and in combination with the CD results described above, tree height can be estimated. Finally, we used the revised CHM data derived from LiDAR to validate the tree height accuracy calculated by the GOMS model.
Results
Tree height and CD results derived from the CHM data
Since there was not enough field-measured tree height data for our study area, CHM data rather than field measurement data were used to provide the tree heights for the inversion results validation process. Local filtering with a canopy height-based variable window size (Popescu et al. 2002) was used to identify a single tree to extract the single-tree height within the super sample plot. The results showed that the field-measured tree height and the extracted single-tree height based on the CHM data have a high correlation, with an R2 equals value of 0.72 (Fig. 6). The CHM data can be used to provide single-tree scale tree height information.
We further set the sampling unit to a size of 25 m × 25 m and extracted all single-tree heights in each sampling unit, then calculated the mean value of all single trees as the mean tree height of this sampling unit. In this section, the function in Fig. 6 was used to revise the CHM data. After the removal of the non-forest pixels based on the supervised classification results indicated in the section of supervised classification results based on the SPOT image, the mean tree height distribution map of the study area at a 25-m spatial resolution was generated, as shown in Fig. 7.
LiDAR data have typically been used to calculate CD in forest studies (Popescu et al. 2011) when field-measured CD values are not available. We constructed the relationship between the tree height derived from the CHM data and the field-measured CD of the super sample plots. The single-tree points in the CHM data with tree height error ranges smaller than 10% compared with field-measured tree heights were selected to build the function shown in Fig. 8. The results showed that tree height derived from the CHM data had a linear relationship with the field-measured CD values, with a high determination coefficient of 0.61. Thus, we used the function in Fig. 8 to calculate the CD (Fig. 9) as a reference for the validation process of the CD results calculated in the section of canopy diameter results estimated by the semi-variance model.
CD derived from the CHM data based on the function indicated in Fig. 8
Canopy diameter results estimated by the semi-variance model
In our study, each of the super sample plots had a size of 25 m × 25 m; therefore, in this part, all the sample plots used to calculate the CD are 25 m × 25 m. The mean field-measured CD of each sample plot can be calculated as follows:
where \( \overline{\mathrm{CD}} \) is the mean CD of the plot, n is the number of trees within the plot, and CDi is the individual tree CD within the stands.
Optimal fitting function between the sill and field-measured canopy diameter
The optimal fitting function between the sill and the field-measured CD is constructed based on the 16 super sample plots. The mean field-measured CD is calculated according to Eq. (18). Because of the low binarization accuracy (e.g., at sample plot No. 14, the quality of the CCD image is low) and insufficient detailed information on the super sample plots (e.g., sample plots No. 4 and No. 5 contained a large hole and presented low LAI with reduced canopy density), only 13 sample plots were selected to provide the CD data for the modeling process. The results in Table 1 show that the ratios (γ25) between the sill under 2-m and 5-m spatial resolution conditions are the most accurate for estimating the CD, with an R2 value of 0. 72 (Fig. 10). The negative correlation (R) in Table 2 indicates that when the spatial resolution of an image decreased, the sill ratio values of larger CD images decreased faster than the smaller CD images; this result also supports the results reported by Song (2007).
Therefore, the optimal fitting function between CD and the sill ratio value was:
Supervised classification results based on the SPOT image
As described in the section of optimal fitting function between the sill and field-measured canopy diameter, the optimal fitting function was built based on the super sample plots sized 25 m × 25 m. To apply Eq. (19) to a larger scale, selected sample plots at a larger scale with areas 25 m × 25 m and high forest vegetation coverage, which are highly similar to the super sample plots, must first be determined. Thus, in this part, a supervised classification process was employed to select the sample plots for further CD calculation at the regional scale. The maximum likelihood method in ENVI was used to perform the supervised classification with SPOT-5 data (described in the section of bidirectional reflectance data and 163 high spatial remote sensing data), and the classification result is shown in Fig. 11 (pixels in green represent the forest coverage zone).
Canopy diameter calculation results for a small experimental plot
We first applied Eq. (19) to a MODIS 500-m pixel within which the super sample plots were located. The small experimental plot information is shown below in Fig. 12.
Based on the classification results (Fig. 12 (upper-right)), we picked out the forest pixels and set the others to black (DN = 0). We next performed a binarization process with the selected forest pixels by setting the sunlit forest crown area to black (DN = 0) and the shaded area to white (DN = 255) (Fig. 12 (bottom-right)). We then divided the binarization results of the small experimental plot into 20 × 20 parts, each sized 25 m × 25 m, and resampled each forest pixel to 2-m and 5-m spatial resolutions to calculate the sill ratio value (γ25) by using the semi-variance model described in the section of semi-variance model. Then, the CD could be estimated for the small experimental plot. The results showed that the threshold of the CD value is from 0 to 4 m, with most values distributed between 2 and 4 m (Fig. 13).
When the CD calculated by the semi-variance model was compared with the CD based on the CHM data for a small experimental plot (the section of Tree height and CD results derived from the CHM data), the difference value (D-value) results (Fig. 14) demonstrated that the difference between the two CD data points was small, with a concentrated distribution from − 1 to 1 m.
We also compared the CD derived from the CCD image of the 13 super sample plots used in Eq. (19) with the CD derived from the CHM data. The results showed that the CDs derived from the CCD image were smaller than those derived from the CHM data, but these values were highly correlated, with an R value of 0.79 and an RMSE of 0.37 m (Fig. 15). The validation results showed that the semi-variance model can be used to precisely calculate CD.
CD calculation results at the regional scale
To expand the CD calculation process to the regional scale, SPOT, CCD and CHM images with the same coverage were used. To precisely match the supervised classification results based on the SPOT image with those of the CCD image used in the semi-variance model and to perform pixel-to-pixel comparisons of the CD data based on the CCD image with the CD data based on the CHM data, these data must be pre-processed. As the spatial resolution of the CHM data is 0.5 m, we first transferred the supervised classification results of both the SPOT data and the CCD image to a spatial resolution of 0.5 m so that the CD calculated based on the CCD image could be compared with the CD derived from the CHM data pixel-to-pixel. In this process, the forest pixels were defined as those with forest vegetation coverage greater than 0.75, which means that in the transferred supervised classification results, the number of forest pixels sized 0.5 m × 0.5 m is more than 187 for the 25 m × 25 m area. Then, the CCD image was used to calculate CD with the supervised classification results at 25-m spatial resolution according to Eq. (19). The calculated CD results shown in Fig. 16 demonstrate that most pixel values were concentrated from 2 to 4 m.
When the CD results shown in Fig. 16 were compared with the CD derived from the CHM data, the D-value results (Fig. 17) showed that most D-values were concentrated from − 1 to 1 m, with RMSE of 1 m for all data. These results showed that this method can precisely calculate CD at the regional scale.
Tree height estimated results based on the GOMS model and the semi-variance model
In this paper, the BRF datasets built as described in the section (Bidirectional reflectance data and high spatial remote sensing data) were at 500-m spatial resolution. Of all the inversion results, the spatial resolution of all the observational data and parameters in the GOMS model was 500 m. However, the spatial resolution of the CD data calculated by the semi-variance model was 25 m (the Section of CD calculation results at the regional scale). When combining the CD data and the GOMS model inversion results to calculate the tree height, we transferred the CD data from 25- to 500-m spatial resolution with Eq. 18. As shown in Fig. 18, the results demonstrated that the standard deviation of a 500-m pixel was mainly concentrated at 0.5 m, which reflected that in one 500 m × 500 m pixel, the difference in CD among the subpixels sized 25 m × 25 m was small. Through this method, high-accuracy CD results at a 500-m spatial resolution could be obtained. Combined with the inversion results (b/R and h/b) provided by the GOMS model, tree height at a 500-m spatial resolution could be calculated (Fig. 19a). We also calculated tree height based on the CHM data at a 500-m spatial resolution by averaging all single trees in one 500-m scale pixel; the results are shown in Fig. 19b.
We compared the inverted tree height calculated by the GOMS model and the tree height derived from the CHM data. In the pixel outlined in black (Fig. 19) wherein the super sample plots were located, the tree height calculated by the GOMS model and that derived from the CHM data were 7.34 and 9.60 m, respectively. The difference between the two tree heights was small.
We also calculated the threshold of the tree height (equals the mean value ± standard deviation value, with the standard deviation value calculated when upscaling CHM tree height from single-tree point to 500-m spatial resolution scale) of each pixel. The relationship between the thresholds and the tree height calculated by the GOMS model is plotted in Fig. 20a. The results showed that, most of the inverted tree heights are within the threshold range, and the GOMS model will overestimate tree height with low tree height pixels. Since the GOMS model was more suitable for a high-FVC coverage area, we then compared the two different tree heights among pixels with high forest coverage ratios (greater than 0.8); the difference between them was small, with an R value of 0.49 and RMSE of 2.44 (Fig. 20b). The low RMSE showed that with CD provided by another data source as prior knowledge for the GOMS model, tree height could be accurately calculated for dense crown areas.
Discussion
The results described in the section of optimal fitting function between the sill and field-measured canopy diameter support the research of Song et al. (2007) with different sample plots of different crown sizes; when the spatial resolution of the sample plots decreased, the sill value decreased faster with larger crown size sample plots than with smaller ones. Moreover, in the optimal fitting function, the sill ratio was closer to the real size of the crown canopy than the spatial resolution of the sample plots.
In the validation process, the data values of the CD (the section of CD calculation results at the regional scale) and inverted tree height (the section of tree height estimated results based on the GOMS model and the semi-variance model) results were close to those of the validation data (with low RMSE), but the R2 was small. We further attempted to determine which factors influenced the accuracy of the inverted tree height. Two reasons were identified, as discussed herein: (1) The inaccuracy of the tree heights derived from the CHM data. In the section of canopy diameter calculation results for a small experimental plot, local filtering with a canopy height-based variable window size was used to extract single-tree points, with an R2 of 0.72 compared with the field-measured tree height. Thus, the extracted tree height used as the true tree height of one pixel could cause errors in the later comparison process. However, due to insufficient field-measured tree height data, these datasets must be used to perform the validation. (2) The unmatched spatial resolution of different datasets may cause errors in the data transference process. As described in the section of canopy diameter results estimated by the semi-variance model, CD results must be based on the transferred CD at 500-m spatial resolution, referring to the effective CD of a 500 m × 500 m pixel, which had a different physical meaning with the CD (true CD) in the GOMS model. This difference may cause an error when calculating tree height using these effective CD data, especially in areas with low FVC.
Since the inaccuracy caused by factor (1) was inevitable due to the shortage of field-measured tree height data, we conducted further testing to explore the importance of the effect of factor (2) on the inaccuracy of the estimated tree height result.
We combined the CD data derived from the CHM data (Fig. 9) with the canopy structural parameters inversion data derived from the GOMS model to calculate tree height and then compared these with the CHM data. The comparison results (Fig. 21) showed that in the pixels with high FVC (forest coverage ratio higher than 0.8), the tree heights derived from the CHM data were slightly higher overall than the tree heights obtained from the GOMS model but also showed high correlation, with an R value of 0.79 and a RMSE of 1.56. The results shown in Fig. 21 were better than those shown in Fig. 20b, which means that when comparing tree height calculated by the GOMS model with that derived from the CHM data, the accuracy uncertainty emerges primarily in the process of transforming CDs to tree heights. However, despite the low consistency, both RMSEs (2.44 and 1.56) shown in Figs. 20b and 21 were low, which demonstrates that the method detailed in our study is suitable for areas with high FVC.
Conclusions
In this study, we provided CD data derived from a high spatial resolution image as a priori knowledge for the GOMS model to obtain tree height data at the regional scale. We first built the optimal relationship function between the sill calculated by the semi-variance model and the field-measured CD, and then applied the optimal function at the regional scale (the section of CD calculation results at the regional scale) to obtain CD data covering the entire image. Moreover, by combing the CD results described in the section of CD calculation results at the regional scale and the canopy structure parameters (b/R and h/b) inversion results derived from the GOMS model, tree height at the regional scale could be obtained (the section of canopy diameter results estimated by the semi-variance model). The results showed that γ25 (the ratio between the sill values when the spatial resolution of the image was 2 and 5 m) had the greatest R2 of 0.72 with the CD. Moreover, the differences between the tree heights calculated by the GOMS model and the tree heights derived from the CHM data were small. We also found that the calculated tree height result had high accuracy in an area with high FVC, exhibiting an RMSE of 2.44 in the pixels for which the forest area coverage index was greater than 0.8.
However, several problems remained unresolved. For example, additional field-measured data are needed for the modeling process to increase the precision of the optimal function when using the semi-variance model to calculate CD. Additionally, many pixels did not match the assumption of the GOMS model; therefore, BRF data at a higher spatial resolution are needed for future studies. Moreover, inconsistencies in the spatial resolution between the calculated CD results and the inversion results of the GOMS model can also lead to inaccurate tree height calculation results. Although there are several uncertainties with the method used in this research, our project provides a novel concept for calculating tree height cheaply and easily, which has far-reaching relevance in the field of forest studies, especially for difficult-to-reach areas or for cases in which prior knowledge of forest structural parameters is lacking. Therefore, in a forthcoming study, we will focus on improving the accuracy of our modeling process and obtain multi-angle BRF data with higher spatial resolution to perform canopy structural parameter inversions.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- GOMS:
-
Geometric-optical mutual shadowing
- CD:
-
Canopy diameter
- CHM:
-
Canopy height model
- DEM:
-
Digital elevation model
- RMSE:
-
Root mean square error
- FVC:
-
Fractional vegetation cover
- AGB:
-
Above-ground biomass
- LAI:
-
Leaf area index
- DBH:
-
Diameter at breast height
- MODIS:
-
Moderate-resolution Imaging Spectro radiometer
- MISR:
-
Multi-angle Imaging Spectro radiometer
- BRF:
-
Bidirectional reflectance
- BRDF:
-
Bidirectional reflectance distribution function
- DN:
-
Digital number
References
Breidenbach J, Næsset E, Lien V, Gobakken T, Solberg S (2010) Prediction of species specific forest inventory attributes using a nonparametric semi-individual tree crown approach based on fused airborne laser scanning and multispectral data. Remote Sens Environ 114(4):911–924
Carmean WH, Lenthall DJ (1989) Height-growth and site-index curves for jack pine in north central Ontario. Can J For Res 19(2):215–224
Chen E, Guo Z (2008) WATER: dataset of forest structure parameter survey at the super site around the Dayekou Guantan forest station. Heihe plan science data center. Institute of Forest Resource Information Techniques, Chinese Academy of Sciences, Heihe Plan Science Data Center, Beijing
Chen HYH, Klinka K, Kabzems RD (1998) Height growth and site index models for trembling aspen (Populus tremuloides Michx.) in northern British Columbia. Forest Ecol Manag 102(2):157–165
Dubayah RO, Sheldon SL, Clark DB, Hofton MA, Blair JB, Hurtt GC, Chazdon RL (2010) Estimation of tropical forest height and biomass dynamics using lidar remote sensing at La Selva, Costa Rica. J Geophys Res Biogeo 115:G00E09
Edson C (2011) Light detection and ranging (LiDAR): what we can and cannot see in the forest for the trees. PhD Dissertation, Oregon State University
Enquist BJ, Brown JH, West GB (1998) Allometric scaling of plant energetics and population density. Nature 395(6698):163–165
Enquist BJ, Niklas KJ (2001) Invariant scaling relations across tree-dominated communities. Nature 410(6829):655–660
Enquist BJ, West GB, Brown JH (2009) Extensions and evaluations of a general quantitative theory of forest structure and dynamics. PNAS 106(17):7046–7051
Ercanlı İ (2020) Innovative deep learning artificial intelligence applications for predicting relationships between individual tree height and diameter at breast height. Forest Ecosyst 7:12
Fu Z, Wang J, Song JL, Zhou HM, Pang Y, Chen BS (2011) Estimation of forest canopy leaf area index using MODIS, MISR, and LiDAR observations. J Appl Remote Sens 5(1):053530
Heinzel J, Koch B (2012) Investigating multiple data sources for tree species classification in temperate forest and use for single tree delineation. Int J Appl Earth Obs 18:101–110
Huo L, Lindberg E (2020) Individual tree detection using template matching of multiple rasters derived from multispectral airborne laser scanning data. Int J Remote Sens 41(24):9525–9544
Jupp DL, Strahler AH, Woodcock CE (1988) Autocorrelation and regularization in digital images. I basic theory. IEEE T Geosci Remote 26(4):463–473
Jupp DL, Strahler AH, Woodcock CE (1989) Autocorrelation and regularization in digital images. II Simple image models. IEEE T Geosci Remote 27(3):247–258
Leckie D, Gougeon F, Hill D, Quinn R, Armstrong L, Shreenan R (2003) Combined high-density lidar and multispectral imagery for individual tree crown analysis. Can J Remote Sens 29(5):633–649
Lefsky MA, Cohen WB, Parker GG, Harding DJ (2002) Lidar remote sensing for ecosystem studies lidar, an emerging remote sensing technology that directly measures the three-dimensional distribution of plant canopies, can accurately estimate vegetation structural attributes and should be of particular intere. Psychol Rep 46(1):927–930
Lefsky MA, Harding DJ, Keller M, Cohen WB, Carabajal CC, Espirito-Santo FDB, Hunter MO, de Oliveira R Jr (2005) Estimates of forest canopy height and above-ground biomass using ICESat. Geophys Res Lett 32:L22S02
Lefsky MA, Keller M, Pang Y, Camargo PBD, Hunter MO (2007) Revised method for forest canopy height estimation from geoscience laser altimeter system waveforms. J Appl Remote Sens 1(1):6656–6659
Li C, Song J, Wang J (2015) Modifying geometric-optical bidirectional reflectance model for direct inversion of forest canopy leaf area index. Remote Sens 7(9):11083–11104
Li X, Gao F, Wang J, Zhu Q (1997) Uncertainty and sensitivity matrix of parameters in inversion of physical BRDF model. J Remote Sens 1(1):5–14
Li X, Liu S, Xiao Q, Ma M, Jin R, Che T, Wang W, Hu X, Xu Z, Wen J, Wang L (2017) A multiscale dataset for understanding complex eco-hydrological processes in a heterogeneous oasis system. Sci Data 4:170083
Li X, Strahler AH (1985) Geometric-optical modeling of a conifer forest canopy. IEEE T Geosci Remote 23(5):705–721
Li X, Strahler AH (1992) Geometric-optical bidirectional reflectance modeling of the discrete crown vegetation canopy: effect of crown shape and mutual shadowing. IEEE T Geosci Remote 30(2):276–292
Li XW, Strahler AH (1986) Geometric-optical bidirectional reflectance modeling of a conifer forest canopy. IEEE T Geosci Remote 24(6):906–919
Li Z, Xu Z (2011) Detection of change points in temperature and precipitation time series in the Heihe River basin over the past 50 years. Resour Sci 33(10):1877–1882
Ma H, Song J, Wang J, Xiao Z, Fu Z (2014) Improvement of spatially continuous forest LAI retrieval by integration of discrete airborne LiDAR and remote sensing multi-angle optical data. Agric For Meteorol 189–190:60–70
Means JE, Acker SA, Harding DJ, Blair JB, Lefsky MA, Cohen WB, Harmon ME, McKee WA (1999) Use of large-footprint scanning airborne lidar to estimate forest stand characteristics in the Western cascades of Oregon. Remote Sens Environ 67(3):298–308
Muller-Landau HC, Condit RS, Harms KE, Marks CO, Thomas SC, Bunyavejchewin S, Chuyong G, Co L, Davies S, Foster R, Gunatilleke S, Gunatilleke N, Hart T, Hubbell SP, Itoh A, Kassim AR, Kenfack D, LaFrankie JV, Lagunzad D, Lee HS, Losos E, Makana J-R, Ohkubo T, Samper C, Sukumar R, Sun I-F, Supardi MNN, Tan S, Thomas D, Thompson J, Valencia R, Vallejo MI, Muñoz GV, Yamakura T, Zimmerman JK, Dattaraja HS, Esufali S, Hall P, He F, Hernandez C, Kiratiprayoon S, Suresh HS, Wills C, Ashton P (2006) Comparing tropical forest tree size distributions with the predictions of metabolic ecology and equilibrium models. Ecol Lett 9(5):589–602
Nelson R, Oderwald R, Gregoire TG (1997) Separating the ground and airborne laser sampling phases to estimate tropical forest basal area, volume, and biomass. Remote Sens Environ 60(3):311–326
Nigh GD, Sit V (1996) Validation of forest height-age models. Can J For Res 26(5):810–818
Nilsson M (1996) Estimation of tree heights and stand volume using an airborne lidar system. Remote Sens Environ 56(1):1–7
Payandeh B (1974) Notes: formulated site index curves for major timber species in Ontario. For Sci 20(2):143–144
Payandeh B, Wang Y (1994a) Modified site index equations for major Canadian timber species. Forest Ecol Manag 64(1):97–101
Payandeh B, Wang Y (1994b) Relative accuracy of a new base-age invariant site index model. For Sci 40(2):341–348
Popescu SC, Wynne RH, Nelson RF (2002) Estimating plot-level tree heights with lidar: local filtering with a canopy-height based variable window size. Comput Electron Agr 37(1):71–95
Popescu SC, Zhao K, Neuenschwander A, Lin C (2011) Satellite lidar vs. small footprint airborne lidar: comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens Environ 115(11):2786–2797
Riaño D, Valladares F, Condés S, Chuvieco E (2004) Estimation of leaf area index and covered ground from airborne laser scanner (Lidar) in two contrasting forests. Agric For Meteorol 124(3–4):269–275
Song C (2007) Estimating tree crown size with spatial information of high resolution optical remotely sensed imagery. Int J Remote Sens 28(15):3305–3322
Song C, Dickinson MB, Su L, Zhang S, Yaussey D (2010) Estimating average tree crown size using spatial information from Ikonos and QuickBird images: across-sensor and across-site comparisons. Remote Sens Environ 114(5):1099–1107
Song C, Woodcock CE (2003) Estimating tree crown size from multiresolution remotely sensed imagery. Photogramm Eng Remote Sens 69(11):1263–1270
Song C, Woodcock CE, Li X (2002) The spectral/temporal manifestation of forest succession in optical imagery: the potential of multitemporal imagery. Remote Sens Environ 82(2):285–302
Sun G, Ranson KJ, Zhang Z (2006) Forest vertical parameters from lidar and multi-angle imaging spectrometer data. J Remote Sens 10(004):523–530
Swatantran A, Dubayah R, Roberts D, Hofton M, Blair JB (2011) Mapping biomass and stress in the Sierra Nevada using lidar and hyperspectral data fusion. Remote Sens Environ 115(11):2917–2930
Thrower JS, Goudie JW (1992) Estimating dominant height and site index of even-aged interior Douglas-fir in British Columbia. West J Appl For 7(1):20–25
Wang G, Klinka K (1995) Site-specific height curves for white spruce (Picea glauca [Moench] Voss) stands based on stem analysis and site classification. Ann For Sci 52(6):607–618
West GB, Enquist BJ, Brown JH (2009) A general quantitative theory of forest structure and dynamics. PNAS 106(17):7040–7045
Xiao Q (2018) HiWATER: Airborne CCD image data in the middle reaches of the Heihe River Basin on July 26, 2012. https://doi.org/10.3972/hiwater.164.2014.db
Xiong BM, Wang ZX, Li ZQ, Zhang E, Tian K, Li TT, Li Z, Song CHL (2016) Study on the correlation among age, DBH and tree height of the Pseudotsuga sinensis in Qizimei Mountain nature reserve. Forest Resour Manage 4:41–46
Yang RC, Kozak A, Smith JHG (1978) The potential of Weibull-type functions as flexible growth curves. Can J For Res 8(4):424–431
Acknowledgements
Sincere thanks to Ma Han for sharing her datasets and method and to the researchers who performed the field-measured work of Dayekou.
Funding
This research was partially supported by the National Natural Science Foundation of China (No. 41871231), and partially supported by the National Key Research and Development Program of China (No. 2016YFB0501502), the Special Funds for Major State Basic Research Project (No. 2013CB733403).
Author information
Authors and Affiliations
Contributions
Congrong Li collected and analyzed the data and wrote the first draft. Jingling Song provided the study idea. Jinling Song and Jindi Wang reviewed the draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, C., Song, J. & Wang, J. New approach to calculating tree height at the regional scale. For. Ecosyst. 8, 24 (2021). https://doi.org/10.1186/s40663-021-00300-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40663-021-00300-4