Abstract
Background
Alfalfa (Medicago sativa L.) is a perennial leguminous forage that can improve the properties of saline soil. In addition, the supplementation with fertilizer to promote soil bacterial activity is critical to improve the productivity of coastal saline soils. However, the association between fertilizer application, bacterial community characteristics, and alfalfa yield in saline soil remains unclear.
Aim
To understand the interaction of different fertilizer and environmental factors on soil bacterial diversity and alfalfa yield in coastal saline soil.
Results
A 4-year field study was conducted to examine the interaction of different fertilizer treatments and environmental factors on soil bacterial diversity and alfalfa yield in coastal saline soil in China. Three organic fertilizer treatments (fulvic acid potassium, fulvic acid potassium + wood vinegar, and fulvic acid potassium + Bacillus), one biochar treatment (bio-charcoal), one inorganic fertilizer treatment (diammonium phosphate), and a control (no fertilizer) were included. The effects of the six treatments and 15 cutting times on alfalfa yield and soil bacterial community diversity were assessed. The productivity in fulvic acid potassium + Bacillus and fulvic acid potassium treatments was 68.37 and 67.90 t·hm−2 in 4 years, which was significantly higher than that in the bio-charcoal treatment and control. Hydrothermal conditions and timing of alfalfa harvest had significant effects on the soil bacterial community. Proteobacteria, Acidobacteria, and Actinobacteria were the principal bacterial phyla detected in the soil samples, collectively accounting for approximately 60% of the total bacterial abundance. The relative abundance of Bacteroidetes, Firmicutes, and Nitrospirae was significantly correlated with alfalfa yield, and the abundance of these phyla was also affected by the maximum temperature and precipitation. Fertilizer supplementation in coastal saline soil can effectively increase the yield of alfalfa. Among the fertilizers tested, fulvic acid potassium had the strongest effect, whereas bio-charcoal fertilizer had little effect on alfalfa yield.
Graphical Abstract

Similar content being viewed by others
Background
Soil salinization, which is the accumulation of water-soluble salts in the soil solum to the extent that agricultural production and environmental health are affected, is of major international concern. Almost 3% of the global soil area is affected by salinization and approximately 3.6% of China’s soil resources are classified as saline-alkali [1, 2]. It is difficult to increase food production to meet the demand from the growing world population by expanding the area of arable land. Improvement of plant productivity in saline-alkali soils is a viable alternative means of attaining food security.
Microbiomes are essential components of soil ecosystems and play a fundamental role in the direct and indirect maintenance of soil fertility and nutrient cycling, and in plant and animal health [3,4,5,6,7]. Introduction of beneficial microorganisms to the soil to promote plant growth and nutrient absorption is an effective strategy to improve land productivity [8, 9]. Bacterial inoculants play an important role in promoting nutrient absorption and photosynthetic activity. For example, Egamberdiyeva [10] observed that maize roots inoculated with Pseudomonas, Bacillus, and Mycobacterium species in nutrient-deficient soil showed enhanced efficiency in the uptake of nutrients (nitrogen, phosphorus, and potassium) than in soil lacking bacterial inoculants. Nutrient availability is crucial for soil microbial communities, especially for bacterial communities [11]. The availability of soil nitrogen and phosphorus affects plant diversity, and bacterial community diversity and structure [12,13,14,15].
Alfalfa (Medicago sativa L.) is an important leguminous forage, which is widely planted in saline soil because of its excellent adaptability, and is considered to increase the soil organic matter content and improve the properties of saline soil [16,17,18]. The higher osmotic potential and unbalanced ion concentration in saline soil severely hinder water and nutrient absorption by plants [19]. Salinization significantly affects the relative abundance of soil bacteria, such as Firmicutes, which increase the incidence of bacterial wilt in potato [20, 21]. A recent experiment on fertilization of saline soil indicated that the bacterial community plays a crucial role in improving the yield of alfalfa [22]. Generally, 10%–40% of the total applied fertilizer is absorbed by plants and the remaining 60%–90% is lost as a result of leaching, denitrification, and volatilization. In this regard, fertilizer application to increase food production is considered environmentally harmful and unsustainable [8, 23]. Microbial inoculants offer a novel approach for improving agricultural productivity and maintaining ecosystem health [24, 25].
The soil microbiome comprises the microbial communities in the soil and their effects on the soil environment [5]. Ecologists are familiar with the dynamics of animal and plant communities, such as their introduction and domestication, pest control, landscape design, grazing, and fishing, but the dynamics of microbial communities are less well understood [26]. Advances in gene-sequencing technologies have helped to enhance our understanding of soil microbial diversity and facilitate the control of targeted microorganisms in soil processes [3, 27]. Although previous studies on the dynamics of microbial communities are limited, a growing body of research has revealed that microbial communities are broadly identifiable, especially those from similar habitats [26, 28,29,30]. Whether the productivity of saline soil can be improved by predicting the dynamic changes in the soil microbial community or by targeting the microbial composition remains uncertain.
A previous meta-analysis of 31 studies on the effects of inorganic and organic fertilizers on soil microbial diversity revealed that, in all studies, the microbial diversity showed high residual heterogeneity, suggesting that detailed investigations were needed to fully understand the effects of fertilization regimes on microbial diversity and ecosystem function [31]. Therefore, in the present study, high-throughput sequencing technology was used to analyze the soil bacterial communities in coastal saline soil treated with different fertilizers. The aim was to screen the most important bacteria that affect alfalfa yield and provide a basis for efficient utilization of soil resources in saline regions.
Materials and methods
Site description and experimental design
The experiment was conducted at Dagang Farm (38°83′ N, 117°45′ E), which is close to Bohai Bay, from May 2016 to September 2019. The area has a temperate monsoon climate, with annual rainfall of 522 mm (primarily distributed between May and September), and evaporation of 2070 mm. The average annual temperature is 14.5℃, and the mean maximum/minimum temperature in the hottest/coldest month is 26.8°C (July)/-3.4°C (January). The growing season is approximately from April to October. The soil type is coastal saline-alkali soil (Entisol in the US soil classification system), the pH is 8.21, salinity is 2.78 g kg−1, organic carbon content is 9.84 g kg−1, available nitrogen is 49.8 mg kg−1, available phosphorus is 6.27 mg kg−1, available potassium is 134.0 mg kg−1, available calcium is 247.3 mg kg−1, and available sodium is 0.892 g kg−1. The experimental plot was established in May 2016 and the sowing rate was 18 kg hm−2. The alfalfa cultivar used in the experiment was ‘Gibraltar’, which has a fall dormancy score of 2 (provided by DLF Seed Industry Co., Ltd.).
The fertilization treatments chosen for this study were: fulvic acid potassium (FAP; mineral origin and fulvic acid ≥ 50%, K2O ≥ 12%), fulvic acid potassium + wood vinegar (FAPWV), fulvic acid potassium + Bacillus megaterium (FAPB), bio-charcoal (BC; derived from bamboo charcoal pyrolysis at 500–600°C), diammonium phosphate (DAP), and no fertilizer (CK). A total of 18 plots were established, comprising 6 fertilization treatments × 3 replications. Each plot was 6 m × 6 m (36 m2). Fertilizer was applied to the soil surface in each spring with irrigation (see Table 1 for details). During the experiment, standard cultivation and management practices were applied, and weeds and insects were removed regularly. Fifteen harvest cuts of alfalfa were performed in July (T1607), August (T1608), and September (T1609) in 2016, in May (T1705), July (T1707), August (T1708) and September (T1709) in 2017, in May (T1805), July (T1807), August (T1808) and September (T1809) in 2018, in May (T1905), July (T1907), August (T1908) and September (T1909) in 2019. The cutting criterion was that approximately 10% of alfalfa was in boom. Detailed weather information for the experiment plot during the study period is presented in Table 2.
Data collection
Prior to cutting, the forage sample composition was determined in a 1 m × 1 m quadrat in each plot with a stubble height of 5 cm. Dry matter yield was determined by drying approximately 500 g of the fresh forage sample at 65 °C for 24 h to a constant weight. Soil samples were collected after the harvest in July, August, and September 2016, and in May and September 2017. A five-point sampling method was used to collect soil cores from each plot, and then the five soil samples were mixed to form one repeat sample. The soil samples were passed through a 1-mm mesh screen and then placed in sterile polyethylene tubes. These tubes were placed in an incubator (filled with liquid nitrogen) and then stored at − 80 °C before DNA extraction.
Soil samples from the 18 plots were collected after the last harvest in September 2019. In each plot, 10 randomly distributed soil cores (to 10 cm depth and 3.2 cm in diameter) were collected with a sampling probe and pooled. The soil samples were sieved through a 2-mm mesh at the sampling site and were used to estimate soil available nitrogen (the sum of extractable soil ammonium (NH4+-N) and nitrate (NO3−-N) concentrations) and available phosphorus contents. Soil NH4+-N was extracted with 1 M KCl, NO3−-N was extracted with 1 M NH4Cl, soil available phosphorus was extracted with 0.5 M NaHCO3, and all parameters were determined with a Complete Soil Kit (SKW500, Palintest). The extractable NH4+-N, NO3−-N, and available phosphorus concentrations were converted to a dry mass basis using soil moisture data.
DNA extraction and PCR amplification
Soil DNA was extracted from 0.25 g of each soil sample using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Then DNA sample was cleaned with the PowerClean® DNA Clean-Up Kit ((MO BIO Laboratories). The DNA concentration and purity were quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
We used the primer set 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′- GGACTACHVGGGTWTCTAAT-3′) to amplify the V3-V4 regions of the bacterial 16S rRNA gene [32]. PCR reactions were performed in triplicate in a 25 μL volume containing 12.5 μL of 2 × Taq PCR MasterMix, 3 μL bovine serum albumin (2 ng uL−1), 1 μL (5 μM) of each primer, and 30 ng of template DNA under the following conditions: pre-denaturation at 95 °C for 5 min, 28 cycles at 95 °C for 45 s, annealing at 55 °C for 50 s and elongation at 72 °C for 45 s with a final extension at 72 °C for 10 min, and holding at 4 °C. After amplification, PCR products were detected in 2% (w/v) agarose gel and the size of each amplicon was not less than 550 bp.
Illumina MiSeq sequencing
The PCR amplicons were extracted from 2% agarose gels and purified with the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), then quantified with QuantiFluor™-ST (Promega, Madison, WI, USA). The purified amplicons were pooled in equimolar amounts, and then were paired-end sequenced (2 × 250) by Allwegene Company (Beijing, China) using the MiSeq PE300 platform (Illumina, San Diego, CA, USA).
Processing of sequencing data
Raw reads were first demultiplexed and quality filtered by USEARCH Version 8.1 (http://www.drive5.com/usearch/) [33], in which the sequences containing fewer than 3 consecutive bases or obtaining a quality score less than 20 were excluded, and sequences with chimeras were also removed. The optimized reads were clustered into operational taxonomic units (OTUs) using UPARSE 7.1 at a threshold of 97% sequence similarity [33]. To compare all samples at the same sequencing level, the minimum reads number was adopted to subsample sequences from all other samples. Subsequently, effective sequences were aligned against the SILVA database (http://www.arb-silva.de). Taxa were identified down to phylum, class, order, family, and genus levels using the Ribosomal Database Project (RDP; http://rdp.cme.msu.edu/) Bayesian classifier at the 70% threshold.
Based on the OTUs information, alpha diversity indices of community diversity (Shannon index), community richness (Chao estimator), and sequencing depth (Good’s Coverage) were calculated with Mothur (http://www.mothur.org/wiki). To detect the potential biomarkers, the linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/) was used based on a normalized relative abundance matrix. The LEfSe method uses the Kruskal–Wallis test to identify features with significant differences between different treatments and performs LDA to evaluate the effect size of each feature [34]. A LDA threshold score of 3 and a significant α of 0.05 were applied for biomarker detection. In addition, the Kruskal–Wallis and Tukey–Kramer tests were used to compare the differences in functional groups among the different treatments with the FAPROTAX database [35].
Statistical analysis
A one-factor analysis of variance (fertilizer or cutting times), as implemented in SPSS 20.0 (SPSS, Inc., Chicago, IL, USA), was conducted to analyze the yield, relative abundance, and alpha diversity data among the treatments. Duncan’s multiple range test was then performed using least significant differences or Tamhane’s T2 post hoc test. The threshold for determining the significant differences was P < 0.05.
Results
Effects of different fertilizers on alfalfa yield and soil nutrition content
The alfalfa yield at each cutting time and the annual yield from 2016 to 2019 are listed in Table 3. A significant difference was detected among the six fertilizer treatments in yield of T1607, total yield of 2016, total yield of 2017, and total yield of 2016–2019, but no significant differences were observed among other cutting times. The maximum yield was observed in the FAP treatment in T1607 (5.17 t hm−2), whereas the BC treatment had the lowest yield (3.70 t hm−2). In 2016, the annual yield of FAPB was highest at 14.73 t·hm−2, whereas the annual yield of the BC treatment was lowest at 10.16 t hm−2. The annual yields in the FAP and FAPB treatments in 2017 were the highest (23.12 and 22.59 t·hm−2, respectively), and were significantly higher than those of the other four treatments. Similarly, the total yield from 2016 to 2019 in the FAP and FAPB treatments was the highest at 67.90 and 68.37 t hm−2, respectively, which represented an increase of 19.7% and 20.5% over the CK (Table 3).
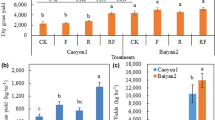
Figure 1 summarizes the effects of the six fertilization treatments on soil available nitrogen and phosphorus contents, as well as the relationship between total yield from 2016 to 2019 and each nutrient. The contents of soil available nitrogen and phosphorus under the FAPB and FAPWV treatments were highest, and were significantly higher than those of the BC, DAP, and CK (Fig. 1a). Pearson correlation analysis revealed that the soil available nitrogen and phosphorus contents were significantly correlated with total yield of 2016–2019 (Fig. 1b and c).
Effects of different fertilizers treatments on soil nitrogen and potassium and their relationship with yield. FAP, fulvic acid potassium; FAPWV, fulvic acid potassium + wood vinegar; FAPB, fulvic acid potassium + Bacillus megaterium; BC, bio-charcoal; DAP, diammonium phosphate; CK, no fertilizer. Different upper-case and lower-case letters above bars in a represent significant differences in available nitrogen and phosphorus contents between fertilizer treatments. The dotted line in b and c represents the 95% confidence interval
Effects of cutting times on soil bacterial community structure
No significant effect on alfalfa yield was observed among the fertilizer treatments (Additional file 1: Fig. S1), but significant differences were detected among the cutting times (Fig. 2a). T1705 had the highest yield at 6.69 t hm−2, which was significantly higher than that of T1608 (4.42 t hm−2), T1607 (4.21 t hm−2), T1609 (3.68 t hm−2), and T1709 (3.14 t hm−2).
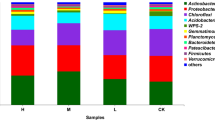
A total of 3,067,467 quality-filtered and chimera-checked 16S rRNA gene sequences were obtained with an average length of 418 bp across all samples. The number of 16S rRNA sequences obtained per sample varied from 17,487 to 161,577. In total, 8824 bacterial OTUs were detected from the 90 DNA samples (Additional file 2). At the phylum level, Proteobacteria, Acidobacteria, and Actinobacteria were the principal bacterial communities detected in the soil samples, and collectively accounted for approximately 60% of the total bacterial abundance (Fig. 2b and Additional file 1: Fig. S1).
PERMANOVA tests confirmed that the bacterial community structures of T1607, T1608, T1609, T1705, and T1709 differed significantly (R2 = 0.519, p = 0.001; Fig. 3). Bacterial diversity index values differed significantly among the five cutting times. The Shannon index indicated that the bacterial community diversity of T1709 was significantly higher than those of the other four fertilizer treatments (Fig. 4a). The Chao estimator indicated that the relative bacterial abundance of T1609 was significantly higher than those of T1608 and T1705 (Fig. 4b).
Nonmetric multidimensional scaling ordination based on the mean abundance value of the bacterial communities in the five cutting time treatments. A permutational multivariate analysis of variance (PERMANOVA) was performed to evaluate the significance of differences in the community structures among the cutting times
Response of bacterial community composition to cutting time
The LEFSe tool was used to identify specialized bacterial communities in the various samples (Fig. 5). Significant indicator groups, determined with the screening criteria LDA (linear discriminant analysis) ≥ 3 and p < 0.05, are shown in Fig. 6. In T1607, only the Proteobacteria phylum (classes Gammaproteobacteria and Alphaproteobacteria) was significantly enriched. In T1608, the phyla Acidobacteria (class Acidobacteria), Bacteroidetes (class Sphingobacteria), and Proteobacteria (class Alphaproteobacteria) were significantly enriched. In T1609, the phylum Proteobacteria (class Deltaproteobacteria) was significantly enriched. In T1705, the phyla Actinobacteria (class Actinobacteria), Bacteroidetes (class Sphingobacteria), and Proteobacteria (classes Alphaproteobacteria and Betaproteobacteria) were significantly enriched. In T1709, three phyla comprising Planctomycetes (class Phycisphaerae), Armatimonadetes, and Parcubacteria were significantly enriched.
Cladogram showing the phylogenetic distribution of the soil bacterial lineages at the five cutting times. Different-colored shading represents different constituents. Rings in this figure indicate phylogenetic levels from phylum (outer) to genus (inner). The diameter of each circle is proportional to the abundance of the group
The relative abundance of bacterial at the phyla at the five cutting times is shown in Fig. 7. At T1607 and T1705, the relative abundance of Actinobacteria, Bacteroidetes, and Firmicutes was highest. The relative abundance of Acidobacteria and Bacteroidetes was highest at T1608, whereas that of Chloroflexi was highest at T1609. For T1709, the relative abundance of Actinobacteria, Chloroflexi, Firmicutes, and Nitrospirae was highest. Pearson correlation analysis indicated that the relative abundance of the above-mentioned seven bacterial phyla was significantly correlated with alfalfa yield (Fig. 7h).
Relative abundance of bacterial phyla at each cutting time (a–g) and their correlation with yield (h). a Acidobacteria, b Actinobacteria, c Bacteroidetes, d Chloroflexi, e Planctomycetes, f Firmicutes, and g Nitrospirae. Bars and error bars in a to g indicate the mean and SE (n = 18). Different lower-case letters above bars indicate a significant difference between cutting times. * p < 0.05, ** p < 0.01, *** p < 0.001
Comparison of the change in relative abundance of the different bacterial phyla with alfalfa yield revealed that the trends for Bacteroidetes, Firmicutes, and Nitrospirae were consistent with alfalfa yield, and that the abundance of these phyla were affected by the maximum temperature and precipitation (Fig. 8). The maximum temperature-promoted increase in yield was associated with positive regulation of Bacteroidetes and Firmicutes, and decreased yield was correlated with negative regulation of Nitrospirae. Precipitation positively regulated the abundance of Bacteroidetes to increase alfalfa yield and negatively regulated Nitrospirae to reduce alfalfa yield.
Pearson correlation analysis of environmental factors, soil bacterial phyla, and alfalfa yield. Tmax, average maximum temperature; Prec, precipitation. Red arrows indicate a significant positive correlation, green arrows indicate a significant negative correlation, and gray arrows indicate a nonsignificant correlation. The values beside the lines are the correlation coefficients. *p < 0.05, **p < 0.01, ***p < 0.001
Comparison of the functional differences of the bacterial communities among the five cutting times resulted in the prediction of 58 significant functional groups (Additional file 2). Based on the differences in alfalfa yield among the treatments, 11 functional groups were consistent with the yield. These functional groups were all associated with nitrogen transformation (Fig. 9).
Discussion
The addition of organic and mineral fertilizers is beneficial to the yield of alfalfa [36,37,38]. Based on a 34-year study, Tang et al. [39] reported that the application of inorganic and organic fertilizers had a strong impact on the soil nitrogen-fixing bacteria community, and that long-term application of organic fertilizer increased the abundance of soil nitrogen-fixing bacteria in paddy fields. Surprisingly, there was no significant differences in soil bacterial diversity were detected among the six fertilizer treatments in the present study. However, fulvic acid potassium effectively increased the yield of alfalfa on saline soil and the additional of wood vinegar would reduced this benefit (Table 3), likely because the activity of certain beneficial microorganisms would be inhibited. Under salt stress, fulvic acid potassium improves the content of neutral protein, and sucrase and urease activities in rhizosphere soil, and increases the abundance of beneficial bacteria (Bacillus) [40], thus improving the soil nutrient status in soil and improving the salt tolerance of plants, but also improves the soluble protein content and antioxidant enzyme activity in cells, thereby reducing the cellular caused by malondialdehyde [41]. No significant difference in yield was observed among treatments in 2018 and 2019 after the discontinuation of fertilizer supplementation (Table 3). The significant differences in alfalfa yield among the cutting times was consistent with the results of Hakl [37], who reported that 80% of the yield difference could be explained by the year. Furthermore, He et al. [22] concluded that organic fertilizer can increase the above-ground net primary production of alfalfa by promoting root growth in saline soil.
Given the impacts on environmental safety and human health caused by traditional inorganic fertilizers, specific soil microbes are under consideration to replace fertilizers as an ecofriendly approach for sustainable agricultural development and food security [42]. Rath et al. [43] showed that the distribution of salt tolerance traits in communities could be quantified by the dose–response relationship between salinity and bacterial growth, which raises the possibility of agricultural utilization of saline soil. Soil microorganisms are beneficial in improving the adaptability of plants in saline soil. For example, Pseudomonas putida improves the absorption rates of K+, Mg2+, and Ca2+, and reduces the absorption of Na+ by cotton grown in high-salinity soil [44, 45]; Achromobacter piechaudii increases the biomass of tomato (Solanum lycopersicum L.) and pepper (Capsicum annuum L.) plants under salt stress [46]. It is noteworthy that the enrichment of soil microorganisms by plants is selective. The present results showed that the relative abundances of Bacteroidetes, Firmicutes, and Nitrospirae were significantly correlated with alfalfa yield (Fig. 6). Although Bacteroidetes are beneficial in promoting the activity of alkaline phosphatase and associated transporters to improve the efficiency of phosphorus acquisition by plants [47, 48], the opposite effect was apparent in the present study. Firmicutes have been shown to enhance tomato wilt and plant growth, but this benefit has not been observed for alfalfa [20, 49, 50]. Huang et al. [51] speculated that reducing the relative abundance of Nitrospirae and increasing the ratio of abundance of Proteobacteria/Acidobacteria in the soil was conducive for improvement of rice yield, which was consistent with the present research. Similarly, Kohler et al. [52] concluded that inoculation with alternative rhizospheric bacteria was an effective means to alleviate salt stress in salt-sensitive plants, which was mainly associated with the increase in activity of antioxidant enzymes induced by beneficial bacteria under severe salt stress. In addition, the production of exopolysaccharides by bacterial populations under adverse environmental conditions has been shown to assist plant roots to absorb water and nutrients by improving the physical properties of the rhizosphere soil, thereby increasing wheat (Triticum aestivum L.) yield [53]. In conclusion, the soil bacterial community plays an important role in plant salt tolerance, but the specific mechanisms and processes involved require further investigation.
Environmental factors profoundly contribute to shaping the soil microbiome, both spatially and temporally [5]. Precipitation, temperature, and their interaction are considered to be important factors affecting grassland microbial diversity [54], and these results were confirmed in the present research (Fig. 7). Yao et al. [55] considered precipitation and temperature to be the key drivers that shape the soil bacterial community in arid and semi-arid systems, and that climatic variables had indirect effects on the soil bacterial community mainly through their direct effects on soil properties (soil nutrients and enzyme activities). However, different ecosystems may support unique microbial populations. For example, precipitation may increase the relative abundance of Bacteroidetes (Fig. 7) [55]. Temperature change limits the ability of microorganisms to survive, disperse, and colonize soil spaces, and alters the soil microbial community structure [56]. Nottingham et al. [57] evaluated the adaptability of a tropical forest soil microbial community to long-term temperature differences and observed that the bacterial community showed growth in the temperature range from − 7.3 to 35 °C. However, bacterial diversity may be reduced when soil is incubated at 35 °C for 112 days [58]. Zogg et al. [59] reported that soil microbial communities differed across a range of temperatures. Some microbiologists hold a different opinion, arguing that microbes are more strongly impacted by plant responses rather than by abiotic manipulation. A previous study evaluated the effects of multi-factor climate change on microbial communities in vegetated and bare plots; the results showed that the bacterial community structure was significantly changed by the treatments in vegetated soils, whereas no differences in the bacterial community across the treatments were observed in bare plots [60].
Conclusions
The present results showed that supplemental fertilizer was indeed beneficial to alfalfa yield and that fulvic acid potassium had the strongest effect on alfalfa yield in coastal saline land, and this advantage was lost after fertilization was stopped. Bio-charcoal fertilizer did not play a role in increasing alfalfa yield, which may be related to its inability to provide nutrients. Significant differences in alfalfa yield were observed among the various cutting times, which were mainly associated with the relative abundance of Bacteroidetes, Firmicutes, and Nitrospirae in the soil. The maximum temperature increased the alfalfa yield by positively regulating the relative abundance of Bacteroidetes and Firmicutes, and negatively regulated the relative abundance of Nitrospirae to reduce the alfalfa yield. Precipitation increased alfalfa yield by positively regulating the relative abundance of Bacteroidetes, and negatively regulated the relative abundance of Nitrospirae to reduce alfalfa yield. The soil bacterial community plays an important role in increasing alfalfa yield in coastal saline soil, among which the microbial functional groups associated with nitrogen transformation are the most important, but the mechanism requires further investigation.
Data availability
The original contribution presented in this study are included in the article/additional material, further inquiries can be directed to the corresponding authors.
References
Rengasamy P. World salinization with emphasis on Australia. J Exp Bot. 2006;57:1017–23. https://doi.org/10.1093/jxb/erj108.
Pan C, Liu C, Zhao H, Wang Y. Changes of soil physico-chemical properties and enzyme activities in relation to grassland salinization. Eur J Soil Biol. 2013;55:13–9. https://doi.org/10.1016/j.ejsobi.2012.09.009.
Bar-On YM, Phillips R, Milo R. The biomass distribution on Earth. Proc Natl Acad Sci U S A. 2018;115:6506–11. https://doi.org/10.1073/pnas.1711842115.
Bender SF, Wagg C, van der Heijden MGA. An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol. 2016;31:440–52. https://doi.org/10.1016/j.tree.2016.02.016.
Islam W, Noman A, Naveed H, Huang Z, Chen HYH. Role of environmental factors in shaping the soil microbiome. Environ Sci Pollut Res Int. 2020;27:41225–47. https://doi.org/10.1007/s11356-020-10471-2.
Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S. The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front Plant Sci. 2017;8:1617. https://doi.org/10.3389/fpls.2017.01617.
Zakavi M, Askari H, Shahrooei M. Bacterial diversity changes in response to an altitudinal gradient in arid and semi-arid regions and their effects on crops growth. Front Microbiol. 2022. https://doi.org/10.3389/fmicb.2022.984925.
Bhardwaj D, Mohammad WA, Ranjan KS, Narendra T. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microbial Cell Fact. 2014;13:66. https://doi.org/10.1186/1475-2859-13-66.
Vinci G, Cozzolino V, Mazzei P, Monda H, Savy D, Drosos M, Piccolo A. Effects of Bacillus amyloliquefaciens and different phosphorus sources on Maize plants as revealed by NMR and GC-MS based metabolomics. Plant Soil. 2018;429:437–50. https://doi.org/10.1007/s11104-018-3701-y.
Egamberdiyeva D. The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl Soil Ecol. 2007;36:184–9. https://doi.org/10.1016/j.apsoil.2007.02.005.
Lakshmanan V, Selvaraj G, Bais HP. Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol. 2014;166:689–700. https://doi.org/10.1104/pp.114.245811.
Beauregard MS, Hamel C, Atul N, St-Arnaud M. Long-term phosphorus fertilization impacts soil fungal and bacterial diversity but not AM Fungal Community in Alfalfa. Microb Ecol. 2009;59:379–89. https://doi.org/10.1007/s00248-009-9583-z.
Clark CM, Cleland EE, Collins SL, Fargione JE, Gough L, Gross KL, Pennings SC, Suding KN, Grace JB. Environmental and plant community determinants of species loss following nitrogen enrichment. Ecol Lett. 2007;10:596–607. https://doi.org/10.1111/j.1461-0248.2007.01053.x.
Yang Z, Hautier Y, Borer ET, Zhang C, Du G. Abundance- and functional-based mechanisms of plant diversity loss with fertilization in the presence and absence of herbivores. Oecologia. 2015;179:261–70. https://doi.org/10.1007/s00442-015-3313-7.
Dang P, Li C, Lu C, Zhang M, Huang T, Wan C, Wang H, Chen Y, Qin X, Liao Y, et al. Effect of fertilizer management on the soil bacterial community in agroecosystems across the globe. Agric Ecosyst Environ. 2022. https://doi.org/10.1016/j.agee.2021.107795.
Bertrand A, Dhont C, Bipfubusa M, Chalifour F-P, Drouin P, Beauchamp CJ. Improving salt stress responses of the symbiosis in alfalfa using salt-tolerant cultivar and rhizobial strain. Appl Soil Ecol. 2015;87:108–17. https://doi.org/10.1016/j.apsoil.2014.11.008.
Song X, Fang C, Yuan Z-Q, Li F-M. Long-term growth of alfalfa increased soil organic matter accumulation and nutrient mineralization in a semi-arid environment. Front Environ Sci. 2021. https://doi.org/10.3389/fenvs.2021.649346.
Yu R, Wang G, Yu X, Li L, Li C, Song Y, Xu Z, Zhang J, Guan C. Assessing alfalfa (Medicago sativa L.) tolerance to salinity at seedling stage and screening of the salinity tolerance traits. Plant Biol Stuttg. 2021;23:664–74. https://doi.org/10.1111/plb.13271.
Muchate NS, Nikalje GC, Rajurkar NS, Suprasanna P, Nikam TD. Plant salt stress: adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot Rev. 2016;82:371–406. https://doi.org/10.1007/s12229-016-9173-y.
Lee SM, Kong HG, Song GC, Ryu CM. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021;15:330–47. https://doi.org/10.1038/s41396-020-00785-x.
Yang C, Wang X, Miao F, Li Z, Tang W, Sun J. Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl Soil Ecol. 2020. https://doi.org/10.1016/j.apsoil.2020.103671.
He F, Wang G, Wang L, Li Z, Tong Z, Wang Y, Li X. Effects of organic base fertilizer and inorganic topdressing on alfalfa productivity and the soil bacterial community in saline soil of the Huanghe River Delta in China. Agronomy. 2022. https://doi.org/10.3390/agronomy12112811.
Adesemoye AO, Kloepper JW. Plant-microbes interactions in enhanced fertilizer-use efficiency. Appl Microbiol Biotechnol. 2009;85:1–12. https://doi.org/10.1007/s00253-009-2196-0.
Shailendra Singh GG. Plant Growth Promoting Rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. J Microbial Biochem Technol. 2015. https://doi.org/10.4172/1948-5948.1000188.
Singh JS, Pandey VC, Singh DP. Efficient soil microorganisms: a new dimension for sustainable agriculture and environmental development. Agr Ecosyst Environ. 2011;140:339–53. https://doi.org/10.1016/j.agee.2011.01.017.
Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013;7:1493–506. https://doi.org/10.1038/ismej.2013.54.
Rout ME. The Plant Microbiome. In: Genomes of Herbaceous Land Plants. 2014. p. 279–309.
Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. https://doi.org/10.1038/ismej.2011.107.
Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. https://doi.org/10.1073/pnas.1000081107.
Shade A, Kent AD, Jones SE, Newton RJ, Triplett EW, McMahon KD. Interannual dynamics and phenology of bacterial communities in a eutrophic lake. Limnol Oceanogr. 2007;52:487–94. https://doi.org/10.4319/lo.2007.52.2.0487.
Bebber DP, Richards VR. A meta-analysis of the effect of organic and mineral fertilizers on soil microbial diversity. Appl Soil Ecol. 2022. https://doi.org/10.1101/2020.10.04.325373.
Yang C, Tang W, Sun J, Guo H, Sun S, Miao F, Yang G, Zhao Y, Wang Z, Sun J. Weeds in the Alfalfa field decrease rhizosphere microbial diversity and association networks in the North China Plain. Front Microbiol. 2022;13: 840774. https://doi.org/10.3389/fmicb.2022.840774.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. https://doi.org/10.1038/ismej.2012.8.
Segata N, Abubucker S, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Waldron L, Zucker J, Thiagarajan M, et al. Microbial community function and biomarker discovery in the human microbiome. Genome Biol. 2011;12:47. https://doi.org/10.1186/gb-2011-12-s1-p47.
McIlroy SJ, Lapidus A, Thomsen TR, Han J, Haynes M, Lobos E, Huntemann M, Pati A, Ivanova NN, Markowitz V, et al. High quality draft genome sequence of Meganema perideroedes str Gr1(T) and a proposal for its reclassification to the family Meganemaceae fam nov. Stand Genomic Sci. 2015;10:23. https://doi.org/10.1186/s40793-015-0013-1.
Lissbrant S, Stratton S, Cunningham SM, Brouder SM, Volenec JJ. Impact of Long-Term Phosphorus and Potassium Fertilization on Alfalfa Nutritive Value-Yield Relationships. Crop Sci. 2009;49:1116–24. https://doi.org/10.2135/cropsci2008.06.0333.
Hakl J, Kunzová E, Konecná J. Impact of long-term organic and mineral fertilization on lucerne forage yield over an 8-year period. Plant Soil Environ. 2016;62:36–41. https://doi.org/10.17221/660/2015-pse.
Hakl J, Kunzová E, Tocauerová Š, Menšík L, Mrázková M, Pozdíšek J. Impact of long-term manure and mineral fertilization on yield and nutritive value of lucerne (Medicago sativa) in relation to changes in canopy structure. Eur J Agron. 2021. https://doi.org/10.1016/j.eja.2020.126219.
Tang H, Li C, Shi L, Xiao X, Cheng K, Wen L, Li W. Effect of different long-term fertilizer managements on soil nitrogen fixing bacteria community in a double-cropping rice paddy field of southern China. PLoS ONE. 2021;16: e0256754. https://doi.org/10.1371/journal.pone.0256754.
Zhang M, Li X, Wang X, Feng J, Zhu S. Potassium fulvic acid alleviates salt stress of citrus by regulating rhizosphere microbial community, osmotic substances and enzyme activities. Front Plant Sci. 2023. https://doi.org/10.3389/fpls.2023.1161469.
Qu Y, Bao G, Pan X, Guo J, Xiang T, Fan X, Zhang X, Yang Y, Yan B, Zhao H, et al. Resistance of highland barley seedlings to alkaline salt and freeze-thaw stress with the addition of potassium fulvic acid. Plant Soil Environ. 2022;68:299–308. https://doi.org/10.17221/84/2022-pse.
Gopal M, Gupta A, Thomas GV. Bespoke microbiome therapy to manage plant diseases. Front Microbiol. 2013;4:355. https://doi.org/10.3389/fmicb.2013.00355.
Rath KM, Fierer N, Murphy DV, Rousk J. Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 2019;13:836–46. https://doi.org/10.1038/s41396-018-0313-8.
Yao L, Wu Z, Zheng Y, Kaleem I, Li C. Growth promotion and protection against salt stress by Pseudomonas putida Rs-198 on cotton. Eur J Soil Biol. 2010;46:49–54. https://doi.org/10.1016/j.ejsobi.2009.11.002.
Kohler J, Caravaca F, Roldán A. An AM fungus and a PGPR intensify the adverse effects of salinity on the stability of rhizosphere soil aggregates of Lactuca sativa. Soil Biol Biochem. 2010;42:429–34. https://doi.org/10.1016/j.soilbio.2009.11.021.
Alavi P, Starcher MR, Zachow C, Muller H, Berg G. Root-microbe systems: the effect and mode of interaction of Stress Protecting Agent (SPA) Stenotrophomonas rhizophila DSM14405(T). Front Plant Sci. 2013;4:141. https://doi.org/10.3389/fpls.2013.00141.
Lidbury I, Borsetto C, Murphy ARJ, Bottrill A, Jones AME, Bending GD, Hammond JP, Chen Y, Wellington EMH, Scanlan DJ. Niche-adaptation in plant-associated Bacteroidetes favours specialisation in organic phosphorus mineralisation. ISME J. 2021;15:1040–55. https://doi.org/10.1038/s41396-020-00829-2.
Fraser T, Lynch DH, Entz MH, Dunfield KE. Linking alkaline phosphatase activity with bacterial phoD gene abundance in soil from a long-term management trial. Geoderma. 2015;257–258:115–22. https://doi.org/10.1016/j.geoderma.2014.10.016.
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Rio TG, Jones CD, Tringe SG, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–4. https://doi.org/10.1126/science.aaa8764.
Qamar N, Rehman Y, Hasnain S. Arsenic-resistant and plant growth-promoting Firmicutes and gamma-Proteobacteria species from industrially polluted irrigation water and corresponding cropland. J Appl Microbiol. 2017;123:748–58. https://doi.org/10.1111/jam.13535.
Huang M, Tian A, Chen J, Cao F, Chen Y, Liu L. Soil bacterial communities in three rice-based cropping systems differing in productivity. Sci Rep. 2020;10:9867. https://doi.org/10.1038/s41598-020-66924-8.
Kohler J, Hernández JA, Caravaca F, Roldán A. Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ Exp Bot. 2009;65:245–52. https://doi.org/10.1016/j.envexpbot.2008.09.008.
Kaci Y, Heyraud A, Barakat M, Heulin T. Isolation and identification of an EPS-producing Rhizobium strain from arid soil (Algeria): characterization of its EPS and the effect of inoculation on wheat rhizosphere soil structure. Res Microbiol. 2005;156:522–31. https://doi.org/10.1016/j.resmic.2005.01.012.
Yang X, Li Y, Niu B, Chen Q, Hu Y, Yang Y, Song L, Wang J, Zhang G. Temperature and precipitation drive elevational patterns of microbial beta diversity in Alpine Grasslands. Microb Ecol. 2021. https://doi.org/10.1007/s00248-021-01901-w.
Yao M, Rui J, Niu H, Heděnec P, Li J, He Z, Wang J, Cao W, Li X. The differentiation of soil bacterial communities along a precipitation and temperature gradient in the eastern Inner Mongolia steppe. CATENA. 2017;152:47–56. https://doi.org/10.1016/j.catena.2017.01.007.
Carson JK, Gonzalez-Quinones V, Murphy DV, Hinz C, Shaw JA, Gleeson DB. Low pore connectivity increases bacterial diversity in soil. Appl Environ Microbiol. 2010;76:3936–42. https://doi.org/10.1128/AEM.03085-09.
Nottingham AT, Baath E, Reischke S, Salinas N, Meir P. Adaptation of soil microbial growth to temperature: Using a tropical elevation gradient to predict future changes. Glob Chang Biol. 2019;25:827–38. https://doi.org/10.1111/gcb.14502.
Lin Y-T, Jia Z, Wang D, Chiu C-Y. Effects of temperature on the composition and diversity of bacterial communities in bamboo soils at different elevations. Biogeosciences. 2017;14:4879–89. https://doi.org/10.5194/bg-14-4879-2017.
Zogg GP, Zak DR, Ringelberg DB, White DC, MacDonald NW, Pregitzer KS. Compositional and functional shifts in microbial communities due to soil warming. Soil Sci Soc Am J. 1997;61:475–81. https://doi.org/10.2136/sssaj1997.03615995006100020015x.
Koyama A, Steinweg JM, Haddix ML, Dukes JS, Wallenstein MD. Soil bacterial community responses to altered precipitation and temperature regimes in an old field grassland are mediated by plants. FEMS Microbiol Ecol. 2018. https://doi.org/10.1093/femsec/fix156.
Acknowledgements
Not applicable.
Funding
This research was supported by the National Natural Science Foundation of China (No. 32071880), the Agricultural Science and Technology Innovation Program (ASTIP-IAS14) and the earmarked fund for CARS (CARS-34). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
JC conceived and designed the experiments, performed the experiments, analyzed the data, prepared the figures and tables, authored or reviewed drafts of the paper, and approved the final draft. ZL conceived and designed the experiments, prepared the figures and tables and reviewed drafts of the paper. FH conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft. ZT and YW conceived and designed the experiments, and approved the final draft. LW, GZ, and YZ performed the experiments, analyzed the data.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Meets ethical standards applicable to the research discipline.
Consent for publication
All authors agree to the publication of the work.
Competing interests
The authors declare there are no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Alfalfa yield and relative abundance of soil bacterial taxa among different fertilizer treatments.
Additional file 2.
Additional data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cao, J., Li, Z., He, F. et al. Organic fertilizer and hydrothermal conditions change the distribution of Medicago sativa L. productivity and soil bacterial diversity in coastal saline soil. Chem. Biol. Technol. Agric. 10, 119 (2023). https://doi.org/10.1186/s40538-023-00490-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-023-00490-9