Abstract
Background
Thioredoxin (Trx) is reduced by thioredoxin reductase. Trx is used in ribonucleoide reduction, assimilatory sulfate reduction, in modulation of protein sulfhydryl groups, and refolding proteins.
Results
A TcTrx (Tc: Taiwanofungus camphorata) cDNA (640 bp, GenBank AY838902.1) encoding a putative thioredoxin (Trx) of 135 amino acid residues with calculated molecular mass of 16.17 kDa was cloned from Taiwanofungus c amphorata. The deduced amino acid sequence containing a motif (Cys-Gly-Pro-Cys) that is highly conserved among the reported Trxs. A three dimensional structural model of the TcTrx has been created based on the known structure of Malassezia sympodialis Trx (MsTrx, PDB ID: 2j23). To characterize the TcTrx, the codon optimized coding region was subcloned into an expression vector and transformed into Saccharomyces cerevisiae. The recombinant His8-tagged TcTrx was expressed and purified by Ni affinity chromatography. The purified enzyme showed a band of approximately 32 kDa (expected dimeric form) on a 12% SDS-PAGE. The molecular mass determined by MALDI-TOF is 33.16 kDa which suggests that the purified enzyme is a dimeric enzyme. Furthermore, the enzyme exhibited TcTrx activity via insulin assay. The Michaelis constant (K M ) value for insulin was 3.78 × 10−2 mM. The enzyme’s half-life of deactivation was 13 min at 45°C. The enzyme was most active at pH 7.
Conclusions
A three dimensional structural model of T. camphorata Trx based on its TcTrx cDNA sequence. The active form of the TcTrx has been successfully expressed in yeast. The enzyme possesses Trx activity and is capable of reduction of disulfide bonds during the formation of newly synthesized proteins.
Similar content being viewed by others
Background
Taiwanofungus camphorata (T. camphorata) is a valued mushroom found only in the forests of Taiwan. It has been used for centuries as health food, among others (Ao et al. [2009]). T. camphorata has been shown to have anti-inflammatory properties (Hsieh et al. [2010]). The active compounds identified from the fruiting bodies of T. camphorata in a submerged culture are benzenoids, diterpenes, maleic and succinic acid derivatives, polysaccharides, steroids, and triterpenoids (Ao et al. [2009]). It can be obtained as health supplements formulated from the mass of T. camphorata from the artificial cultivation by an Asian Nova Biotechnology Inc (http://www.asian-bio.com/) at a high market value. Many studies were aimed to find the exact bioactive compounds of the mushroom (Ao et al. [2009]). In order to search for physiologically active components including antioxidant enzymes, we have established an expressed sequence tag (EST) from the fruiting bodies of T. camphorata. Based on the established EST, several antioxidant enzymes including a 2-Cys peroxiredoxin (Huang et al. [2007]), a catalase (Ken et al. [2008]), a dithiol glutaredoxin (Ken et al. [2009]), a 2-Cys peroxiredoxin isozyme (Liau et al. [2010]), a monothiol glutaredoxin (Ken et al. [2011]), a nitroreductase (Chen et al. [2012]), a peroxiredoxin (Huang et al. [2014]) and an aryl-alcohol dehydrogenase (Ken et al. [2014]) have been cloned and characterized. This encourages us further to search for active components from the established EST of T. camphorata for potential health food applications.
Thioredoxin (Trx) is reduced by thioredoxin reductase with NADPH as the hydrogen donor. Trx is used in refolding proteins ((Li and Churchich [1997]), in modulation of protein sulfhydryl groups (Buchanan et al. [1994]), and phage assembly (Russel and Mode [1986]). Here, we report a three dimensional structural model of Taiwanofungus camphorata thioredoxin based on its sequence. The coding sequence of the TcTrx cDNA was introduced into an S. cerevisiae expression system and the active enzyme purified and characterized its properties.
Methods
Isolation of TcTrx cDNA
We have previously established an EST database from fruiting bodies of T. camphorata and sequenced all clones with insert size greater than 0.4 kb (data not shown). The identity of a Trx cDNA clone was assigned by comparing the inferred amino acid sequence in various databases using the basic local alignment search tool (BLAST) (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The Trx cDNA fragment was subcloned into pCR®4-TOPO® (invitrogen, CA) vector and transformed into E. coli TOPO10. The nucleotide sequence of the insert was determined in both strands. Sequence analysis revealed that the Trx cDNA covered an open reading frame of a putative Trx cDNA (640 bp, GenBank AY838902.1).
Bioinformatics analysis of TcTrx
The BLAST program was used to search homologous protein sequences in the nonredundant database (NRDB) at the National Center for Biotechnology Information, National Institutes of Health (http://www.ncbi.nlm.nih.gov/). Multiple alignments were constructed using ClustalW2 program. Protein secondary structure was predicted by SWISS-MODEL program and represented as α helices and β strands. A three dimensional structural model of TcTrx was created by SWISS-MODEL (Arnold et al. [2006]) (http://swissmodel.expasy.org/) based on the known crystal structure of Malassezia sympodialis Trx (MsTrx, PDB ID: 2j23).
Subcloning of TcTrx cDNA into an expression vector
TcTrx cDNA was subcloned into an E. coli and yeast expression vector, respectively. The coding region of the TcTrx cDNA was amplified using gene specific flanking primers. The 5′ upstream primer contains Eco RI recognition site (5′GAA TTC GAT GTT ATC TTC GCT TGC ATC C3′) and the 3′ downstream primer contains Eco RI recognition site (5′GAA TTC GCG AGG CCC TGG ATG AG3′). Using 0.2 μg of TcTrx cDNA as a template, and 10 pmole of each 5′ upstream and 3′ downstream primers, a 405 bp fragment encoding the putative mature TcTrx gene was amplified by PCR. The fragment was ligated into pCRÒ4-TOPOÒ and transformed into E. coli. The recombinant plasmid was isolated and digested with Eco RI. The digestion products were separated on a 1.0% agarose gel. The 405 bp insert DNA was gel purified and subcloned into Eco RI site of pET-20b(+) expression vector (Novagen, Darmstadt, Germany). The recombinant DNA was then transformed into E. coli C43(DE3). However, the recombinant protein was not expressed in the E. coli expression system. We then subcloned the gene into the Eco RI site of the pYEX-S1 expression vector (Clontech, Mountain View, CA, USA) and introduced into Saccharomyces cerevisiae (trp− ura−), The recombinant protein was still not expressed in the Saccharomyces cerevisiae expression system. We decided to optimize the TcTrx DNA sequenced based on the yeast codon usage table (The codons were optimized by using the Codon Optimization Tool provided by the Integrated DNA Technologies (http://sg.idtdna.com/CodonOpt) with codon usage table of Saccharomyces cerevisiae. The GenScript Codon Usage Frequency Table Tool was used as the reference for yeast usage table. The optimized gene was custom synthesized by Genomics company, Taiwan. The optimized sequence is shown in Figure 1 in red. It was subcloned into a pET-20b(+) expression vector and the recombinant DNA transformed into E. coli C43(DE3). The recombinant protein was still not expressed in the E. coli expression system. We then re-amplified the codon-optimized pET-20b(+)-TcTrx DNA using two gene-specific primers: the 5′ upstream primer contained Eco RI recognition site (5′GAA TTC GAT GTT ATC TTC GCT TGC ATC C3′) and the 3′ downstream primer contained a His8-tag and Eco RI recognition site (5′ GAATTC GAG ACG TCA GTG GTG GTG GTG GTG GTG GTG GTG3′). Using the 0.2 μg optimized recombinant DNA of pET-20b(+)-TcTrx as a template, and 10 pmole of each 5′ upstream and 3′ downstream primers, a 0.4 kb fragment was amplified by PCR. The fragment was ligated into pCRÒ4-TOPOÒ and transformed into E. coli. The recombinant plasmid was isolated and digested with Eco RI. The digestion products were separated on 1.0% agarose gel. The 0.4 kb insert DNA was gel purified and subcloned into the Eco RI site of the pYEX-S1 expression vector and introduced into Saccharomyces cerevisiae (trp− ura−). The transformed yeast cells were selected by YNBDT (0.17% yeast nitrogen base, 0.5% ammonium sulfate, and 2% glucose) agar plates containing 20 μg Trp/mL. The presence of TcTrx cDNA in the selected transformants was verified by PCR using gene-specific flanking primers. The recombinant TcTrx protein was expressed in yeast in YPD medium (1% yeast extract, 2% peptone, 2% glucose). Expression of the functional recombinant TcTrx was analyzed by enzyme activity assay.
Expression and purification of the recombinant TcTrx
The yeast transformant containing the TcTrx gene was grown at 30°C, 170 rpm in 100 mL of YPD medium for 18 h. The cells were harvested and the soluble proteins extracted in PBS (phosphate buffer saline) with glass beads as described previously (Ken et al. [2005]). The recombinant TcTrx was purified by Ni-NTA affinity chromatography (elution buffer: 1× PBS/5% glycerol/1 mM DTT containing 20–250 mM imidazole) according to manufacturer’s instruction (Qiagen). The purified protein was analyzed by a 12% SDS-PAGE followed by staining with Coomassie Brilliant Blue R-250 and destaining with 10% acetic acid/10% methanol. The purified protein was pooled, desalted, and buffer exchanged using Amicon ultra centrifugal filter unit (5000 MWCO). The exchanged buffer exchanged was 0.01 × PBS/0.01 mM DTT/2.5 mM imidazole/0.1% glycerol. The final recombinant TcTrx protein concentration was determined by a Bio-Rad Protein Assay Kit (Richmond, CA). For storage, one volume of glycerol was added to the purified protein and stored at −20°C for further analysis.
Molecular mass analysis via JOEL MALDI-TOF
The purified recombinant TcTrx (0.82 μg/μL) was prepared in 0.01 × PBS containing 0.01 mM DTT, 2.5 mM imidazole and 0.1% glycerol. The sample (5 μL) was used for molecular mass determination using an MALDI-TOF mass spectrometer (JMS-S3000, Japan).
TcTrx activity assay
Trx activity was assayed by the method of Holmgren (Holmgren and Reichard [1967]; Holmgren [1979a], [b]) using the insulin precipitation assay which was monitored by a spectrophotometric assay of the increase in turbidity at 650 nm. The reaction mixture (200 μL) at 25°C contained 0.1 M potassium phosphate (pH 7.0), 2 mM EDTA, 0.33 mM DTT, 15 mM NADPH, 0.025 mM insulin and 1.0 μg TcTrxR (thioredoxin reductase from T. camphorata, Huang et al. [2010]). The reaction was started by the addition of 1.0 μg TcTrx (0.41 μg/μL). The reaction was followed by the increase in A 650 due to insulin precipitation on reduction.
Kinetic studies
The kinetic properties of the TcTrx (1.0 μg) was determined by varying the concentrations of insulin (0.025 ~ 0.055 mM). The change in absorbance at 650 nm was recorded between 30 and 60 sec. The KM, Vmax and kcat were calculated from Lineweaver-Burk plots.
Enzyme characterization
The TcTrx enzyme was tested for stability in terms of its activity under various conditions. Aliquots of the TcTrx sample (1.0 μg) were treated as follows: (1) Thermal effect. Each enzyme sample (1.0 μg) was heated at 40, 50, or 60°C for 5 min. Then samples were checked for TcTrx activity: 80% residual activity at 40°C treatment, 40% residual activity at 50°C treatment. Therefore, we choose at 45°C heating to this enzyme effect, each enzyme sample (1.0 μg) was heated at 45°C for 2, 4, 8, 16 min. (2) pH effect. Each enzyme sample (1.0 μg) was adjusted to desired pH by adding a half volume of buffer with different pHs: 0.2 M citrate buffer (pH 4.0), 0.2 M phosphate buffer (pH 6.0, 7.0 or 8.0) or 0.2 M CAPS buffer (pH 10.0). Each sample was incubated at 37°C for 30 min. At the end of each treatment, samples were checked for TcTrx activity by insulin precipitation assay at pH 7.
Results and discussion
A three dimensional structural model of Taiwanofungus camphorata thioredoxin based on its TcTrx cDNA sequence
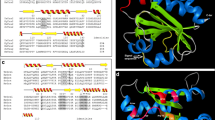
A putative TcTrx cDNA (640 bp) clone was identified on the basis of the consensus pattern and sequence homology to other published Trxs in NCBI database. The entire coding region of TcTrx cDNA is 405 bp and the deduced protein consists of 135 amino acid residues with a calculated molecular mass of 16.2 kDa (accession no. GenBank AY838902.1). The TcTrx gene was optimized based on yeast codon usage table as shown Figure 1 red color codon. Figure 2 shows the optimal alignment of the amino acid sequences of the TcTrx with five selected Trx sequences from other sources. This TcTrx shared 63% similarity with TvTrx (Trametes versicolor, accession no. EIW63845.1), 62% similarity with DsTrx (Dichomitus squalens, accession no. EJF67406.1), 58% similarity with PsTrx (Punctularia strigosozonata, accession no. EIN14269.1), 57% similarity with MsTrx (Malassezia sympodialis, accession no. AJ937746.1, PDB ID: 2j23), and 42% similarity with SmTrx (Schistosoma mansoni, accession no. 2XBI_A). The deduced amino acid sequence containing highly conserved active site motif (Cys62-Gly-Pro-Cys65) (Limacher, et al. [2007]). The two Cys residues are the key to the ability of thioredoxin to reduce other proteins. A three dimensional structural model of the TcTrx (purple) has been created based on the known structure of Malassezia sympodialis Trx (MsTrx, PDB ID: 2j23). The RMSD is 0.95 Å. The highly conserved active motif is denoted in yellow underline (Figure 2A). The secondary structure, predicted by SWISS-MODEL program, showed 4 α helices and 6 β strands. Superimposition with SmTrx (PDB ID: 2XBI, white) was shown using protein solid ribbons. Putative active residues are shown in yellow (Figure 2B).
Alignment of the amino acid sequences of TcTrx with other organism’s Trx and three dimensional structural model. (A) Sequence alignment: TcTrx (this study), TvTrx (Trametes versicolor, EIW63845.1), DsTrx (Dichomitus squalens, EJF67406.1), PsTrx (Punctularia strigosozonata, EIN14269.1), MsTrx (Malassezia sympodialis, AJ937746.1), SmTrx (Schistosoma mansoni, 2XBI_A). Identical amino acids in all sequences are shaded black, conservative replacements are shaded gray. The highly conserved motif is denoted in yellow underline (Figure 1A) predicted by SWISS-MODEL program and represented as α helices and β strands. (B) A three dimensional structural model of TcTrx. The structural model of the TcTrx was created based on the known crystal structure of Malassezia sympodialis (MsTrx, PDB ID: 2j23) via SWISS-MODEL program. Superimposition of TcTrx (purple) and SmTrx (white) was shown using protein solid ribbons. Yellow structure denotes active site.
Expression and purification of the recombinant optimized TcTrx
The optimized coding region for the TcTrx (405 bp) was amplified by PCR and subcloned into an expression vector, pYEX-S1 as described in the Materials and Methods. Positive clones were verified by DNA sequence analysis. The recombinant TcTrx was expressed, and the proteins were analyzed by a 12% SDS-PAGE in the absence of reducing agent and without boiling (Figure 3). The recombinant TcTrx was expressed as a His8-tagged fusion protein and was purified by affinity chromatography with nickel chelating Sepharose. A major band with molecular mass of ~35 kDa (expected size of TcTrx dimer) was detected in Ni-NTA eluted fractions by SDS-PAGE (Figure 3, lanes 8–9). Analysis of the TcTrx by MALDI-TOF MS confirms the presence of a single protein with molecular mass of 33154 Da. This indicates that the enzyme is predominantly dimeric in nature. As shown in Figure 2, from the point of the structure that Cys62 and Cys65, we assume that the Trx favors formation of dimers through intermolecular disulfide linkages. This assumption is supported by our SDS-PAGE analysis of the purified protein in the absence of β-mercaptoethanol, the dimeric form is predominant (see Figure 3, lanes 8–9). The dimers may be linked by two disulfide bonds between Cys62-Cys65 and Cys65-Cys62, or between Cys62-Cys62 and Cys65-Cys65. The dimers may also be linked by one disulphide bond between Cys62 and Cys65, or Cys62-Cys62, or Cys65-Cys65 (Chae et al. [1994]; Liau et al. [2010]). The yield of the purified His8-tagged TcTrx was 82 μg from 100 mL of culture. Functional TcTrx was detected by activity assay as describe below.
Expression and purification of recombinant TcTrx in Saccharomyces cerevisiae. Fifteen μL (loading buffer without mercaptoethanol and without boiling) of each fraction was loaded into each lane of the 12% SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining. Lane 1, crude extract from Saccharomyces cerevisiae expressing TcTrx; 2, flow-through proteins from the Ni-NTA column; 3, wash; 4–9, TcTrx eluted from Ni-NTA column. Molecular masses (in kDa) of standards are shown at left.
Kinetic studies of the purified TcTrx
As shown in Figure 4, the Lineweaver-Burk plot of the velocity (1/V) against 1/[insulin] gave the KM, kcat and kcat/KM values were 3.7×10−2 mM, 1.2×10−2 min−1, 3.1×101 min−1 mM−1. Comparison of the KM with that of Trx from other sources (Table 1) reveals that T. camphorata’s KM is several fold larger. The result indicating that the TcTrx works at higher substrate concentration. But the kcat value is similar to that of Trx from other sources.
Properties of the purified TcTrx
The TcTrx enzyme was shown to possess Trx activity by its ability to reduction insulin. Heat stability of the TcTrx was tested to examine the effect of heat on the Trx activity as described in the Materials and Methods. We found 80% and 40% residual activity when the enzyme was treated at 40°C and 50°C for 5 min, respectively. The enzyme activity found at 25°C was defined as 100% activity. The enzyme (1.0 μg) was further heated for various time intervals at 45°C. The enzyme activity decreased as the heating time increased (Figure 5). The enzyme’s half-life of deactivation was 13 min at 45°C. In Figure 6, the TcTrx is most activity under pH 7.0.
Conclusion
A three dimensional structural model of T. camphorata Trx based on its TcTrx cDNA sequence. This study reported the first cloning and expression of an important reduction enzyme, TcTrx, from T. camphorata. The active form of the TcTrx has been successfully expressed in yeast. The enzyme possesses Trx activity and is capable of reduction of disulfide bonds during the formation of newly synthesized proteins.
The TcTrx has a higher KM value (Table 1) therefore can work under higher substrate concentration. Its kcat value is compatible to that of Trx from another source. It is likely that the Trx is one of the enzymes responsible for reducing protein disulfide targets in T. camphorata.
Abbreviations
- Trx:
-
Thioredoxin
- IPTG:
-
Isopropyl β-D-thiogalactopyranoside
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PBS:
-
Phosphate buffer saline
References
Ao ZH, Xu ZH, Lu ZM, Xu HY, Zhang XM, Dou WF: Niuchangchih ( Antrodia camphorata ) and its potential in treating liver diseases. J Ethnopharmacol 2009, 121: 194–212. 10.1016/j.jep.2008.10.039
Arnold K, Bordoli L, Kopp J, Schwede T: The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 2006, 22: 195–201. 10.1093/bioinformatics/bti770
Buchanan BB, Schurmann P, Decottignies P, Lozano RM: Thioredoxin: a multifunctional regulatory protein with a bright future in technology and medicine. Arch Biochem Biophy 1994, 314: 257–260. 10.1006/abbi.1994.1439
Chae HZ, Uhm TB, Rhee SG: Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc Natl Acad Sci U S A 1994, 91: 7022–7026. 10.1073/pnas.91.15.7022
Chen CC, Ken CF, Wen L, Chang CF, Lin CT: Taiwanofungus camphorata nitroreductase: cDNA cloning and biochemical characterization. Food Chem 2012, 135: 2708–2713. 10.1016/j.foodchem.2012.07.024
Holmgren A: Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested functions of thioredoxin in mechanism of hormone action. J Biol Chem 1979, 254: 9113–9119.
Holmgren A: Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 1979, 254: 9627–9632.
Holmgren A, Reichard P: Thioredoxin 2: Cleavage with cyanogen bromide. Eur J Biochem 1967, 2: 187–196. 10.1111/j.1432-1033.1967.tb00125.x
Hsieh YH, Chu FH, Wang YS, Chien SC, Chang ST, Shaw JF, Chen CY, Hsiao WW, Kuo YH, Wang SY: Antrocamphin A, an anti-inflammatory principal from the fruiting body of Taiwanofungus camphoratus , and its mechanisms. J Agric Food Chem 2010, 58: 3153–3158. 10.1021/jf903638p
Huang JK, Ken CF, Huang HM, Lin CT: Biochemical characterization of a novel 2-Cys peroxiredoxin from Antrodia camphorata . Appl Microbiol Biotechnol 2007, 74: 84–92. 10.1007/s00253-006-0626-9
Huang CY, Chen YT, Wen L, Sheu DC, Lin CT: A peroxiredoxin cDNA from Taiwanofungus camphorata : role of Cys31 in dimerization. Mol Biol Rep 2014, 41: 155–164. 10.1007/s11033-013-2848-0
Ken CF, Hsiung TM, Huang ZX, Juang RH, Lin CT: Characterization of Fe/Mn-superoxide dismutase from diatom Thallassiosira weissflogii : cloning, expression, and property. J Agric Food Chem 2005, 53: 1470–1474. 10.1021/jf048269f
Ken CF, Chen HT, Chang RC, Lin CT: Biochemical characterization of a catalase from Antrodia camphorata : Expression in Escherichia coli and enzyme properties. Bot Stud 2008, 49: 119–125.
Ken CF, Lin CY, Jiang YC, Wen L, Lin CT: Cloning, expression and characterization of an enzyme possessing both glutaredoxin and dehydroascorbate reductase activity from Taiwanofungus camphorata . J Agric Food Chem 2009, 57: 10357–10362. 10.1021/jf9021256
Ken CF, Chen IJ, Lin CT, Liu SM, Wen L, Lin CT: Monothiol glutaredoxin cDNA from Taiwanofungus camphorata : A novel CGFS-type glutaredoxin possessing glutathione reductase activity. J Agric Food Chem 2011, 59: 3828–3835. 10.1021/jf1048113
Ken CF, Chang CC, Wen L, Huang JK, Lin CT: Biochemical characterization of a functional recombinant aryl-alcohol dehydrogenase from Taiwanofungus camphorata . Bot Stud 2014, 55(14):2–10.
Li W, Churchich JE: Activation of partially folded mitochondrial malate dehydrogenase by thioredoxin. Eur J Biochem 1997, 246: 127–132. 10.1111/j.1432-1033.1997.t01-1-00127.x
Liau YJ, Chen YT, Lin CY, Huang JK, Lin CT: Characterisation of 2-Cys peroxiredoxin isozyme (Prx1) from T aiwanofungus camphorata (Niu-chang-chih): Expression and enzyme properties. Food Chem 2010, 119: 154–160. 10.1016/j.foodchem.2009.06.008
Limacher A, Glaser AG, Meier C, Schmid-Grendelmeier P, Zeller S, Scapozza L, Crameri R: Cross-reactivity and 1.4-Å crystal structure of Malassezia sympodialis thioredoxin (Mala s 13), a member of a new pan-allergen family. J Immunol 2007, 178: 389–396. 10.4049/jimmunol.178.1.389
Obiero J, Pittet V, Bonderoff SA, Sanders DAR: Thioredoxin system from Deinococcus radiodurans . J Bacteriol 2010, 192: 494–501. 10.1128/JB.01046-09
Pieulle L, Stocker P, Vinay M, Nouailler M, Vita N, Brasseur GI, Garcin E, Sebban-Kreuzer C, Dolla A: Study of the thiol/disulfide redox systems of the Anaerobe Desulfovibrio vulgaris points out pyruvate: ferredoxin oxidoreductase as a new target for thioredoxin 1. J Biol Chem 2011, 286: 7812–7821. 10.1074/jbc.M110.197988
Russel M, Mode P: The role of thioredoxin in filamentous phage assembly. Construction, isolation, and characterization of mutant thioredoxins. J Biol Chem 1986, 261: 14997–15005.
Acknowledgements
This work was supported by the National Science Council of the Republic of China, Taiwan under grant MOST 103-2313-B-019 -006 to C-T. Lin.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
P-FH and Y-TC carried out the molecular genetic studies and biochemical studies. C-TL was an adviser to carry out the molecular genetic and biochemical studies. LW and Y-TC participated in drafting the manuscript. C-TL and LW drafted the manuscript and revised the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, YT., Hong, PF., Wen, L. et al. Molecular cloning and characterization of a thioredoxin from Taiwanofungus camphorata. Bot Stud 55, 77 (2014). https://doi.org/10.1186/s40529-014-0077-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40529-014-0077-z