Abstract
We identified an autosomal dominant progranulin mutation carrier without symptoms of dementia in her lifetime (Reduced Penetrance Mutation Carrier, RedPenMC). This resistance to develop expected pathology presents a unique opportunity to interrogate neurodegenerative mechanisms. We performed multimodal single-nuclei analyses of post-mortem frontal cortex from RedPenMC, including transcriptomics and global levels of chromatin marks. RedPenMC had an increased ratio of GRN-expressing microglia, higher levels of activating histone mark H3k4me3 in microglia and lower levels of the repressive chromatin marks H3k9me1 and H3k9me3 in the frontal cortex than her affected mutation carrier son and evidence of higher protein levels of progranulin in both plasma and brain homogenates. Although the study is limited to one case, the results support that restoring brain progranulin levels may be sufficient to escape neurodegeneration and FTD. In addition to previously identified modifier genes, it is possible that epigenetic marks may contribute to the increased progranulin expression in cases of reduced penetrance. These findings may stimulate similar follow-up studies and new therapeutic approaches.
Similar content being viewed by others
Introduction
In 2006, it was demonstrated that pathogenic progranulin (GRN) mutations can cause autosomal dominant Frontotemporal dementia (FTD), histopathologically characterised by aggregation of ubiquitin-binding protein p62 and phosphorylated TAR DNA-binding protein 43 (pTDP-43) in the frontal cortex [4, 11, 15]. GRN is a pleiotropic growth factor and pathogenic mutations reduce GRN protein levels in the brain, plasma and cerebrospinal fluid [11]. Various studies identified modifiers of disease risk and age at onset in GRN mutation carriers [27, 28, 42, 65, 71]. In rare cases, mutation carriers completely escape the disease phenotype known as “reduced penetrance” [2, 12]. The underlying mechanism behind reduced penetrance is unclear but may be influenced by several factors including transcriptomic and epigenetic modifications as well as other factors [12]. There are distinct changes in the transcriptome and chromatin landscape in different brain regions of dementia when compared to aged controls [5, 7, 26, 58, 69]. Growing evidence is also showing a link between aging and changes in histone profiles, both the number of histone proteins per cell and in the balance of activating and repressing histone modifications [7, 10, 52].
Lately, high-throughput, droplet-based single-nuclei RNA-Seq (snRNA-Seq) has been employed to study different neurological disorders [20, 35, 50, 62]. Moreover, snRNA-Seq has been combined with nuclei hashing using oligo-barcoded antibodies to allow for multiplexing without altering transcriptional profiles [18]. Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq) is a multimodal method that enables simultaneous analysis of transcriptome and protein targets at a single-cell resolution [55].
This study includes a rare case of reduced penetrance (RedPenMC), a 96-year-old female GRN mutation (p.Tyr294*) carrier who had no signs of FTD in her lifetime, her affected GRN mutation carrier (AMC) son from the same family and an unaffected non-carrier (NC). We used the frontal cortex to perform single-cell multimodal measurements [72] of the transcriptome and global levels of several histone modifications using CITE-Seq [18] since bulk tissue transcriptome analysis will be inadequate to map cell-type-specific molecular changes [62, 72].
Case presentation
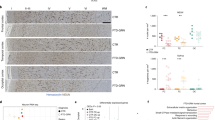
A 96-year-old female (RedPenMC), carrying the pathogenic GRN mutation (p.Tyr294*) [11] and who was devoid of any cognitive impairment or neurological abnormalities until death was identified (Fig. 1A, B). Our Sanger sequencing of cDNA (synthesized from total RNA) from frozen prefrontal cortex Brodmann Area 10 (BA10) confirmed that mutant mRNA was not expressed in RedPenMC (Fig. 1C), which is in line with previous data that GRN non-sense mutations cause non-sense mediated mRNA decay, leading to haploinsufficiency [53]. The GRN haploinsufficiency of the affected mutation carrier (AMC.26) as well as the RedPenMC, was confirmed by reduced serum GRN levels (Fig. 1D). The Genetic Frontotemporal Dementia Initiative (GENFI) have reported that plasma levels below 61.55 ng/mL is a predictor of GRN mutations [16] and more recently the suggested threshold was 71.0 ng/mL [51]. The relative GRN level was higher in RedPenMC (45.70 ng/ml) compared to AMC.26 (31.49 ng/ml). Whole-genome sequencing (WGS) of blood DNA was used to scan for genetic modifiers associated with GRN mediated FTD in RedPenMC, and two of her offspring AMC.26, and the control NC.94 (Additional file 2: Table S5). The genotypes for rs5848, a GRN 3’ UTR variant, was (C/T) in AMC.26 and (C/C) in RedPenMC. The ‘C’ allele is a less efficient binding site for miR-659 compared to the ‘T’ allele, where miR-659 is suggested to reduce GRN expression by inhibiting its translation [23]. Another reported modifier known to affect GRN levels as well as the age of onset [13, 14, 17, 61] in GRN mediated FTD is SNP rs1990622 in the TMEM106B gene, implicated in the proper functioning of the lysosome. The A/G genotype (G protective allele) was present in RedPenMC whereas the A/A genotype (A risk allele) was present in AMC.26. Genotypes of other possible modifier genes (GFAR2, CDH23, PSAP, CELSR2, SORT1) are presented in Additional file 2: Table S5. RedPenMC carries only one copy of the rs7869 T-allele associated with increased PSAP and lower plasma PGRN levels [37], no C-allele for rs646776 which is associated with increased SORT1 mRNA levels and reduced plasma PGRN levels [9] and finally RedPenMC is homozygous AA for GFAR2 where AA homozygotes have an increased disease risk associated with decreased brain GFRA2 mRNA levels but no reported effect on extracellular levels of PGRN [42].
Clinical, genetic and immunohistochemical analysis of AMC and RedPenMC. A Pedigree of the RedPenMC (I-2). 96-year-old female with GRN (p.Tyr294*) mutation. AOD: age of death, AO: age of symptom onset. Also see Additional file 2: Table S1. B Genotyping by Sanger sequencing of the family members for the GRN (p.tyr294*) mutation showing the T > G substitution (c.882T > G) at g.42428777T > G known as rs794729670 [11, 39]. C Sanger sequencing of GRN (exon9) cDNA generated from frozen brain BA10 of RedPenMC showing only the wild type allele T at position c.882. D Serum levels of GRN measured using ELISA in three technical replicates for each donor. E Immunohistochemical staining of the frontal cortex of AMC.26 (red arrows) and RedPenMC against p409/410 TDP-43 and p62. A 96-yr-old female NC was used as a control. Hematoxylin was used as a nuclear counterstain. The control paraffin sections were obtained from the Netherlands Brain Bank (NBB, refer Additional file 2: Table S2b). Scale bar:200 µm & 2 µm. L2/3: Cortical layer 2/3
Immunohistochemistry in RedPenMC showed an absence of immunoreactivity towards p62 and pTDP-43 in the frontal cortex similar to the findings in age and gender-matched NC whereas AMC.26 showed characteristic p62 and pTDP-43 aggregations (Fig. 1E, red arrows).
Single-cell multimodal analysis of RedPenMC, AMC.26 and NC.38
CITE-Seq was performed to comprehend cell-type-specific molecular changes in RedPenMC and AMC.26 (Fig. 2A–D, details refer Additional files 1 & 2). Frozen BA10 from RedPenMC, from an unrelated age and gender-matched control (NC.38) and from AMC.26 (Additional file 2: Table S2a) were processed for unbiased isolation of nuclei and the nuclei were stained for a panel of 15 nuclear proteins (Additional file 2: Table S3) including histone marks followed by sequencing using 10X genomics chromium. After quality control, we obtained 18,266 single nuclear transcriptome profiles from RedPenMC, AMC.26 and NC.38 with a median detection of 2223 expressed genes per cell. In parallel, nuclei hashing yielded 9925 protein profiles after the quality control, we used the Nuclear Pore Complex (NPC) for quantification. Cell clusters were identified using the Seurat single cell analysis toolkit [56], and the distinct cell clusters were annotated according to different cell lineage markers, resulting in the detection of 15 cell types including excitatory and inhibitory cortical neurons, astrocytes and microglia (Fig. 2B, C). Neuronal subtypes included excitatory neurons upper layer CUX2, middle layer RORB, deep layer TSHZ2 [59] as well as inhibitory neurons (IN) marked with GAD1, which included different subtypes IN-PV, IN-SST, IN-VIP and IN-SV2C (Fig. 2C). All the cell types were represented in all three samples except IN-SST (only in NC.38), T-cells (only in AMC.26 and RedPenMC) and perivascular macrophages (only in RedPenMC). These cell types were not considered in the subsequent analysis. Using cellular annotation from the transcriptome, we mapped the global levels of 14 nuclear proteins for each cell type (Fig. 2D). The protein panel included chromatin marks: H3k4me3, H3k9me1 and H3k9me3.
Overview of the experimental approach and CITE-Seq datasets. A Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq) multi-modal analysis of nuclei isolated from frozen Brodmann area 10 (BA10) which includes NC.38, AMC.26 and reduced penetrance (RedPenMC, refer Additional file 2: Table S1 & S2a). NC.38 frozen brain was obtained from Mount Sinai Brain bank part of NIH Brain and Tissue Repository (NIH NeuroBioBank, USA). AMC.26 and RedPenMC frozen brain were obtained from Brain Bank at Karolinska Institutet, Sweden. B UMAP of integrated snRNA-Seq 18,266 profiles from AMC.26, NC.38 and RedPenMC annotated according to the expression of known markers. C Known markers for pan-neuronal, excitatory neurons, glial and endothelial cells (OPCs: Oligodendrocyte progenitor cells). D Representative protein profiles for different cell types after normalization of the unique molecular identifier (UMI) counts, (ADT Antibody derived tags)
We assessed the GRN expression by analysing the expression levels of the different GRN-splice variants in frozen BA10 (refer Additional file1 & 2) from RedPenMC, AMC (n = 3), and unrelated NC (age and gender-matched to AMC and RedPenMC, n = 3, Fig. 3B). Overall, the total GRN expression, including all splice variants (S1, S2, S3, S4) was higher (P < 0.005) in RedPenMC compared to AMC. Especially, splice variant ‘S2’ was significantly increased in RedPenMC compared to AMC and NC. The ‘S2’ splice variant upregulated in RedPenMC, includes rs5848 in the 3’ UTR region [44]. These results indicate that total GRN expression in bulk RNA was higher in RedPenMC compared to AMC and NC, at least for some isoforms.
Fraction of GRN expressing cells in the BA10 region and cell-type-specific associated changes in RedPenMC and AMC. A The ratio between number of GRN expressing to non-expressing cells for each cell type in Brodmann area 10 (BA10) sampled from RedPenMC, AMC.26 and NC.38. The P value calculated by comparing different cell types. *indicates P < 0.05. B Different GRN splice variants (S1, S2, S3, S4) expressed in BA10 region of NC (n = 3), AMC (n = 3) and RedPenMC (n = 1) analyzed using Digital Droplet™ PCR (ddPCR). RNA was isolated from the bulk tissue for each donor. GRN splice variants were detected amplifying exon-exon junctions, for S1:E1_E2, S2:E13_3’UTR, S3:E6_E7 and S4:E10_E12. Transcript expression is expressed as number of copies per microliter. The threshold was set automatically above any positive signal detected in multiples negative controls that contained a digital droplet PCR (ddPCR) cocktail and water in place of either of the cDNA. Multiple negative controls were included in each plate. Statistical differences in the expression of four splice variants in NC, AMC and RedPenMC were calculated by one-way ANOVA with Bonferroni's multiple comparison (Post Hoc test). *P < 0.05, ****P < 0.005. C Differentially expressed genes (DEGs) in RedPenMC, AMC.26 and NC.38 (upregulated and downregulated) obtained from all cell types. P values were calculated by comparing the number of DEGs between different cell types per group comparison. *indicates P < 0.05. D The intersection of DEGs in different comparison groups
GRN is a ubiquitously expressed protein [36]. To characterize the GRN expression in BA10 of RedPenMC, we computed the distribution of cells expressing detectable levels of GRN across each cell type (Fig. 3A). The fraction of microglia expressing GRN was higher in RedPenMC (P < 0.05) compared to AMC.26 and NC.38 (Fig. 3A). However, there was only a trend towards a significantly lower total GRN expression in AMC.26 compared to RedPenMC (P = 0.054, log2fold change − 0.254), possibly due to the low sequencing-depth and relatively low overall signal to noise GRN expression levels. We also observed a high concordance between GRN expression and PSAP expression [71] in microglia in both GRN mutation carriers (Additional file 2: Table S4). Western blot analysis of homogenized frozen BA10 showed that RedPenMC had brain progranulin protein-levels closer to the levels in the control NC.38 than the levels in AMC.26 (Additional file 3: Fig. S2A).
To apprehend the transcriptome profile of RedPenMC, we compared nuclear profiles with AMC.26 and NC.38 (Additional file 2: Table S6). We detected 465 differentially expressed genes (DEGs, P < 0.05; log2fold change ± 0.5) in the comparison between RedPenMC and AMC.26 (Fig. 3C) and 555 DEGs between AMC.26 and NC.38. Finally, we compared RedPenMC and NC.38, resulting in the detection of 589 DEGs.
In RedPenMC vs AMC.26, the highest number of DEGs were found in microglia followed by astrocytes and endothelial cells. The statistical testing revealed the DEGs burden in microglia were significant (P < 0.05) compared to other cell types. Similar DEGs burden analysis per cell type was carried out for AMC.26 vs NC.38 and RedPenMC vs NC.38 and the analysis yielded no significant differences (Fig. 3C). Previously, it was reported that GRN haploinsufficiency can be extended to the microglial expression of GRN [20, 49] and loss of GRN affects microglial phenotype as well as transcriptome profile. Next, we explored the intersection of DEGs in microglia and 257 Disease-associated microglial genes (DAM) [8, 29, 35, 54] and found APBB2, PMP22, CD9 to be upregulated in the AMC.26 (Additional file 2: Table S6). GO analysis of DEGs indicated that “astrocyte differentiation”, “oligodendrocyte differentiation” and “regulation of neuron differentiation” were among the enriched dysregulated pathways (Additional file 3: Fig. S1A). Thereafter, we intersected the number of DEGs and GO among three comparison groups described earlier (Fig. 3D, Additional file 3: Fig. S1B). 10% of DEGs were overlapping between RedPenMC vs AMC.26 and AMC.26 vs NC.38. Moreover, 26% of DEGs were overlapping between RedPenMC vs NC.38 and AMC.26 vs NC.38 (Fig. 3D). GO intersection indicates that RedPenMC vs AMC.26 and AMC.26 vs NC.38 share 21% of biological processes (Additional file 3: Fig. S1B). In contrast, GO-intersection analysis of RedPenMC vs NC.38 did not show overlap with any of the other two comparisons (Additional file 3: Fig. S1B). The results may suggest that the identified GO in our study were associated to the pathological phenotype in affected mutation carrier and not the mutation per se or a combination of the mutation and pathological phenotype.
Finally, we performed analysis of global changes of open and closed chromatin marks to understand the chromatin state of RedPenMC and AMC.26 across different cell types. These analyses revealed that 9 out of 14 nuclear proteins (Additional file 2: Table S7) were changed (P < 0.05) among different cell types in AMC.26, RedPenMC and NC.38 (Fig. 4A, B, 5A).
Cell-type-specific changes of histone marks in the BA10 region in RedPenMC and AMC. Global expression profile of 5 distinct nuclear proteins in different cell types among RedPenMC, AMC.26 and NC.38 in neuronal cell types (A) and non-neuronal cell types (B). The p values for the donor versus donor comparison were listed in the Additional file 2: Table S7. The boxplot represent variation among the cells of each cell type in each donor
Cell-type-specific changes of H2A2 in the BA10 region in RedPenMC and AMC. A Global expression profile of H2A2 in different cell types among RedPenMC, AMC.26 and NC.38. The p values for the donor versus donor comparison were listed in the Additional file 2: Table 7. The boxplot represent variation among the cells of each cell type in each donor
In cortical layer 2/3 (L2/3), NeuN expression was reduced in AMC.26 compared with RedPenMC and NC.38 (Fig. 4A). Typically, in FTD associated with GRN mutation, cortical layers 2/3 are affected by the loss of neurons [30] and downregulation of NeuN levels [67]. NeuN quantitative reduction in L2/3 is indicative of a decline in neuronal health in the cortex of AMC.26, our results are in agreement with previous findings of FTD mediated by GRN mutations [30, 67].
The repressive mark H3k9me1 and H3k9me3 were overexpressed in AMC.26 compared to RedPenMC and NC.38 across different cell types analysed (Fig. 4A-B). H3k9me1 is a key mark for functional heterochromatin [47, 48] and preferred substrate for suppressor of variegation 3–9 (SUV39) methyltransferase, which catalyses H3k9me3 formation [33, 48]. Elevated levels of H3k9me1 in mouse models of Alzheimer's disease (AD) [63] leads to the downregulation of BDNF (brain-derived neurotrophic factor). BDNF is critical for neuronal synaptic plasticity and mediates neuronal differentiation and cognitive functions [40, 46]. Moreover, GRN is co-transported with BDNF within neuronal axon and dendrites [41]. H3k9me3 is associated with transcriptional silencing and altered chromatin plasticity in neurodegeneration [32, 66]. Moreover, H3k9me3 is essential in cell identity establishment and maintenance [36]. Previously, overexpression of H3k9me3 has been reported in AD [32] and Huntington disease (HD) [31]. RedPenMC showed a higher expression of H2A2 than AMC.26 in L2/3, L4, astrocytes, microglia, oligodendrocytes, endothelial cells and OPCs (Fig. 5A). H2A2 is critical in stabilizing differentiated cell identity as well as chromatin compaction [34, 57].
On the other hand, global levels of H3k4me3 were decreased in both RedPenMC and AMC.26 across cell types compared to NC.38 (Fig. 4A, B). However, microglial H3k4me3 expression was higher (P < 0.05) in RedPenMC compared to AMC.26. H3k4me3 levels are associated with the upregulation of transcription activity, reflects the extent of transcription and ‘activating’ histones [6, 22]. Recently, Proximity Ligation-Assisted ChIP-Seq (PLAC-Seq) analysis indicated that GRN expression is regulated by H3k4me3 in microglia [38]. Previously, reduced levels of H3k4me3 have been reported in HD, and locus-specific loss and gain of H3k4me3 have been implicated in AD [7, 19].
Conclusions and discussion
Here, we investigated a GRN mutation carrier with reduced penetrance using WGS and multi-modal genomics single-cell analyses [49]. Previously, TMEM106B [15] and GFRA2 [42] were identified as modifiers of GRN mediated FTD. As of date, we lack a systematic understanding of the disease process and modifiers [65]. Reduced penetrance cases have the potential to uncover endogenous escape mechanisms [2, 12]; providing insights that may be useful for designing future molecular therapies. Naturally occurring cases of reduced penetrance can only be explored using patient-derived materials and cannot be replaced by genome-editing or mouse models [2, 12].
Our bulk ddPCR and Western blot analyses revealed a higher brain progranulin RNA and protein expression in RedPenMC compared to AMC. Our single-cell analyses revealed an increased ratio of GRN expressing microglia as well as microglial-specific higher levels of H3k4me3 and lower levels of global H3k9me1 and H3k9me3 expression in RedPenMC compared to AMC. RedPenMC carried one or more extra copies of the “protective” genetic-modifier-variants in GRN, TMEM106B, SORT1 and PSAP in contrast to her affected son. Although our study is limited by sample size and there is a great need of finding a larger number of RedPenMC for example by collaborative sharing of patient materials or screening genomic population genomic data, such as the SweGen [1], we still think our data can generate some interesting new ideas hypothesis and caveats are presented below.
For GRN haploinsufficiency, the obvious therapeutic approach is to increase the progranulin protein levels. However, it is unclear whether an increase of GRN in all cell types, or in specific cell types are needed to maintain normal brain function. Our data suggests that selective increase of GRN expression in microglia, with drug modulation [60] or genome engineering [3, 45] may prevent the initiation of the neurodegenerative process which has successfully been demonstrated in mouse models earlier [68]. In addition to that, combinatorial inhibition of G9a, SETDB1 and GLP may contribute to the modulation of histone methylation enzymes; which has been proven effective in models of AD [70] and HD [31]. In addition, the levels of H3k4me3 can be increased by inhibiting the activity of histone deacetylases [25, 64]. However, future studies are needed in RedPenMC to understand the microglial cellular processing of GRN into granulin peptides [24] and its interaction with neuronal and astrocyte counterparts.
The CITE-Seq approach is limited in resolution and by technical artefacts, including doublets and difficulties in assembling the transcripts. These technical limitations could be overcome through cell sorting and running full-transcript length single-cell sequencing [21] on similar materials, allowing the pinpointing of specific transcripts (alternative splice forms) in different cell populations. Alternatively, microglia could be sorted from BA10 and then sequenced using long-read sequencing. Genome-wide single-cell chromatin accessibility [43] analysis of BA10 is needed to understand cell-type-specific H3k9me3 and H3k9me1 binding sites in RedPenMC, AMC and NC. Although the two CITE-sequenced mutation carriers included were biological relatives and obtained via the same brain biobank, the control was obtained from another biobank, was unrelated and matched with respect to age and sex only to RedPenMC which may further limit the interpretation of the data.
To conclude, our preliminary analysis of a single reduced penetrance case indicates that the presence of sustainable levels of both RNA and protein progranulin, at least in some cell types in brain, may be the result of both genetic as well as epigenetic modifiers. It is possible that a near normal level of progranulin is sufficient to preserve cognition and prevent neurodegeneration but needs to be further explored and we hope our data can generate new testable hypothesis for therapeutic intervention in presymptomatic GRN carriers.
Data availability
CITE-Seq datasets are available at the ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) with accession ID: E-MTAB-9530.
References
Ameur A, Dahlberg J, Olason P, Vezzi F, Karlsson R, Martin M, Viklund J, Kähäri AK, Lundin P, Che H et al (2017) SweGen: a whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur J Hum Genet 25:1253–1260. https://doi.org/10.1038/ejhg.2017.130
Arboleda-Velasquez JF, Lopera F, O’Hare M, Delgado-Tirado S, Marino C, Chmielewska N, Saez-Torres KL, Amarnani D, Schultz AP, Sperling RA et al (2019) Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med 25:1680–1683. https://doi.org/10.1038/s41591-019-0611-3
Arrant AE, Filiano AJ, Unger DE, Young AH, Roberson ED (2017) Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain 140:1447–1465. https://doi.org/10.1093/brain/awx060%JBrain
Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S et al (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442:916–919. https://doi.org/10.1038/nature05016
Belzil VV, Katzman RB, Petrucelli L (2016) ALS and FTD: an epigenetic perspective. Acta Neuropathol 132:487–502. https://doi.org/10.1007/s00401-016-1587-4
Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E et al (2014) H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158:673–688. https://doi.org/10.1016/j.cell.2014.06.027
Berson A, Nativio R, Berger SL, Bonini NM (2018) Epigenetic regulation in neurodegenerative diseases. Trends Neurosci 41:587–598. https://doi.org/10.1016/j.tins.2018.05.005
Butovsky O, Weiner HL (2018) Microglial signatures and their role in health and disease. Nat Rev Neurosci 19:622–635. https://doi.org/10.1038/s41583-018-0057-5
Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, Hunter TA, DeJesus-Hernandez M, Bisceglio GD, Mackenzie IR et al (2010) Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet 87:890–897. https://doi.org/10.1016/j.ajhg.2010.11.002
Cheung P, Vallania F, Warsinske HC, Donato M, Schaffert S, Chang SE, Dvorak M, Dekker CL, Davis MM, Utz PJ et al (2018) Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell 173:1385-1397.e1314. https://doi.org/10.1016/j.cell.2018.03.079
Chiang HH, Forsell C, Lilius L, Oijerstedt L, Thordardottir S, Shanmugarajan K, Westerlund M, Nennesmo I, Thonberg H, Graff C (2013) Novel progranulin mutations with reduced serum-progranulin levels in frontotemporal lobar degeneration. Eur J Hum Genet 21:1260–1265. https://doi.org/10.1038/ejhg.2013.37
Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H (2013) Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet 132:1077–1130. https://doi.org/10.1007/s00439-013-1331-2
Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC et al (2011) Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol 68:581–586. https://doi.org/10.1001/archneurol.2010.350
Feng T, Mai S, Roscoe JM, Sheng RR, Ullah M, Zhang J, Katz II, Yu H, Xiong W, Hu F Loss of TMEM106B and PGRN leads to severe lysosomal abnormalities and neurodegeneration in mice. 50:219. Doi https://doi.org/10.15252/embr.202050219
Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L et al (2011) TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 76:467–474. https://doi.org/10.1212/WNL.0b013e31820a0e3b
Galimberti D, Fumagalli GG, Fenoglio C, Cioffi SMG, Arighi A, Serpente M, Borroni B, Padovani A, Tagliavini F, Masellis M et al (2018) Progranulin plasma levels predict the presence of GRN mutations in asymptomatic subjects and do not correlate with brain atrophy: results from the GENFI study. Neurobiol Aging 62:245.e249-245.e212. https://doi.org/10.1016/j.neurobiolaging.2017.10.016
Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL, Chesi A, Manduchi E, Wells AD, Grant SFA et al (2017) A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet 101:643–663. https://doi.org/10.1016/j.ajhg.2017.09.004
Gaublomme JT, Li B, McCabe C, Knecht A, Yang YM, Drokhlyansky E, Van Wittenberghe N, Waldman J, Dionne D, Nguyen L et al (2019) Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat Commun. https://doi.org/10.1038/s41467-019-10756-2
Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M (2015) Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 518:365–369. https://doi.org/10.1038/nature14252
Grubman A, Chew G, Ouyang JF, Sun G, Choo XY, McLean C, Simmons RK, Buckberry S, Vargas-Landin DB, Poppe D et al (2019) A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat Neurosci 22:2087–2097. https://doi.org/10.1038/s41593-019-0539-4
Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks GJ, Larsson AJM, Faridani OR, Sandberg R (2020) Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol 38:708–714. https://doi.org/10.1038/s41587-020-0497-0
Howe FS, Fischl H, Murray SC, Mellor J (2017) Is H3K4me3 instructive for transcription activation? BioEssays 39:1–12. https://doi.org/10.1002/bies.201600095
Hsiung GY, Fok A, Feldman HH, Rademakers R, Mackenzie IR (2011) rs5848 polymorphism and serum progranulin level. J Neurol Sci 300:28–32. https://doi.org/10.1016/j.jns.2010.10.009
Kao AW, Mckay A, Singh PP, Brunet A, Huang EJ (2017) Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci 18:325–333. https://doi.org/10.1038/nrn.2017.36
Kerimoglu C, Sakib MS, Jain G, Benito E, Burkhardt S, Capece V, Kaurani L, Halder R, Agís-Balboa RC, Stilling R et al (2017) KMT2A and KMT2B mediate memory function by affecting distinct genomic regions. Cell Rep 20:538–548. https://doi.org/10.1016/j.celrep.2017.06.072
Klein HU, McCabe C, Gjoneska E, Sullivan SE, Kaskow BJ, Tang A, Smith RV, Xu J, Pfenning AR, Bernstein BE et al (2019) Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat Neurosci 22:37–46. https://doi.org/10.1038/s41593-018-0291-1
Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, Lam TT, Strittmatter SM (2017) Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 95:281-296.e286. https://doi.org/10.1016/j.neuron.2017.06.026
Kleinberger G, Capell A, Haass C, Van Broeckhoven C (2013) Mechanisms of granulin deficiency: lessons from cellular and animal models. Mol Neurobiol 47:337–360. https://doi.org/10.1007/s12035-012-8380-8
Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z et al (2017) The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47:566-581.e569. https://doi.org/10.1016/j.immuni.2017.08.008
Lee EB, Porta S, Michael Baer G, Xu Y, Suh E, Kwong LK, Elman L, Grossman M, Lee VMY, Irwin DJ et al (2017) Expansion of the classification of FTLD-TDP: distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol 134:65–78. https://doi.org/10.1007/s00401-017-1679-9
Lee J, Hwang YJ, Kim Y, Lee MY, Hyeon SJ, Lee S, Kim DH, Jang SJ, Im H, Min SJ et al (2017) Remodeling of heterochromatin structure slows neuropathological progression and prolongs survival in an animal model of Huntington’s disease. Acta Neuropathol 134:729–748. https://doi.org/10.1007/s00401-017-1732-8
Lee MY, Lee J, Hyeon SJ, Cho H, Hwang YJ, Shin JY, McKee AC, Kowall NW, Kim JI, Stein TD et al (2020) Epigenome signatures landscaped by histone H3K9me3 are associated with the synaptic dysfunction in Alzheimer’s disease. Aging Cell 19:e13153. https://doi.org/10.1111/acel.13153
Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G (2009) The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. Embo Rep 10:769–775. https://doi.org/10.1038/embor.2009.90
Martire S, Banaszynski LA (2020) The roles of histone variants in fine-tuning chromatin organization and function. Nat Rev Mol Cell Biol. https://doi.org/10.1038/s41580-020-0262-8
Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X et al (2019) Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570:332–337. https://doi.org/10.1038/s41586-019-1195-2
Nicetto D, Zaret KS (2019) Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr Opin Genet Dev 55:1–10. https://doi.org/10.1016/j.gde.2019.04.013
Nicholson AM, Finch NA, Almeida M, Perkerson RB, van Blitterswijk M, Wojtas A, Cenik B, Rotondo S, Inskeep V, Almasy L et al (2016) Prosaposin is a regulator of progranulin levels and oligomerization. Nat Commun 7:11992. https://doi.org/10.1038/ncomms11992
Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, Han CZ, Pena M, Xiao J, Wu Y et al (2019) Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366:1134–1139. https://doi.org/10.1126/science.aay0793
Oijerstedt L, Chiang HH, Bjorkstrom J, Forsell C, Lilius L, Lindstrom AK, Thonberg H, Graff C (2019) Confirmation of high frequency of C9orf72 mutations in patients with frontotemporal dementia from Sweden. Neurobiol Aging. https://doi.org/10.1016/j.neurobiolaging.2019.03.009
Park H, Poo MM (2013) Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci 14:7–23. https://doi.org/10.1038/nrn3379
Petoukhov E, Fernando S, Mills F, Shivji F, Hunter D, Krieger C, Silverman MA, Bamji SX (2013) Activity-dependent secretion of progranulin from synapses. J Cell Sci 126:5412–5421. https://doi.org/10.1242/jcs.132076
Pottier C, Zhou X, Perkerson RB 3rd, Baker M, Jenkins GD, Serie DJ, Ghidoni R, Benussi L, Binetti G, Lopez de Munain A et al (2018) Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol 17:548–558. https://doi.org/10.1016/s1474-4422(18)30126-1
Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE et al (2018) Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci 21:432–439. https://doi.org/10.1038/s41593-018-0079-3
Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ, Josephs KA et al (2008) Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet 17:3631–3642. https://doi.org/10.1093/hmg/ddn257
Raitano S, Ordovàs L, De Muynck L, Guo W, Espuny-Camacho I, Geraerts M, Khurana S, Vanuytsel K, Tóth BI, Voets T et al (2015) Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Rep 4:16–24. https://doi.org/10.1016/j.stemcr.2014.12.001
Rauti R, Cellot G, D’Andrea P, Colliva A, Scaini D, Tongiorgi E, Ballerini L (2020) BDNF impact on synaptic dynamics: extra or intracellular long-term release differently regulates cultured hippocampal synapses. Mol Brain. https://doi.org/10.1186/s13041-020-00582-9
Rivera C, Gurard-Levin ZA, Almouzni G, Loyola A (2014) Histone lysine methylation and chromatin replication. Biochim Biophys Acta 1839:1433–1439. https://doi.org/10.1016/j.bbagrm.2014.03.009
Rivera C, Saavedra F, Alvarez F, Díaz-Celis C, Ugalde V, Li J, Forné I, Gurard-Levin ZA, Almouzni G, Imhof A et al (2015) Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res 43:9097–9106. https://doi.org/10.1093/nar/gkv929
Schier AF (2020) Single-cell biology: beyond the sum of its parts. Nat Methods 17:17–20. https://doi.org/10.1038/s41592-019-0693-3
Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, Vistnes S, Stockley JH, Young A, Steindel M et al (2019) Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573:75–82. https://doi.org/10.1038/s41586-019-1404-z
Sellami L, Rucheton B, Ben Younes I, Camuzat A, Saracino D, Rinaldi D, Epelbaum S, Azuar C, Levy R, Auriacombe S et al (2020) Plasma progranulin levels for frontotemporal dementia in clinical practice: a 10-year French experience. Neurobiol Aging 91:167.e161-167.e169. https://doi.org/10.1016/j.neurobiolaging.2020.02.014
Sen P, Shah PP, Nativio R, Berger SL (2016) Epigenetic Mechanisms of Longevity and Aging. Cell 166:822–839. https://doi.org/10.1016/j.cell.2016.07.050
Skoglund L, Brundin R, Olofsson T, Kalimo H, Ingvast S, Blom ES, Giedraitis V, Ingelsson M, Lannfelt L, Basun H et al (2009) Frontotemporal dementia in a large Swedish family is caused by a progranulin null mutation. Neurogenetics 10:27–34. https://doi.org/10.1007/s10048-008-0155-z
Sobue A, Komine O, Hara Y, Endo F, Mizoguchi H, Watanabe S, Murayama S, Saito T, Saido TC, Sahara N et al (2021) Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol Commun 9:1. https://doi.org/10.1186/s40478-020-01099-x
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14:865. https://doi.org/10.1038/nmeth.4380
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single-cell data. Cell 177:1888-1902.e1821. https://doi.org/10.1016/j.cell.2019.05.031
Sun Z, Bernstein E (2019) Histone variant macroH2A: from chromatin deposition to molecular function. Essays Biochem 63:59–74. https://doi.org/10.1042/ebc20180062
Swarup V, Hinz FI, Rexach JE, Noguchi KI, Toyoshiba H, Oda A, Hirai K, Sarkar A, Seyfried NT, Cheng C et al (2019) Identification of evolutionarily conserved gene networks mediating neurodegenerative dementia. Nat Med 25:152–164. https://doi.org/10.1038/s41591-018-0223-3
Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S et al (2018) Shared and distinct transcriptomic cell types across neocortical areas. Nature 563:72–78. https://doi.org/10.1038/s41586-018-0654-5
Telpoukhovskaia MA, Liu K, Sayed FA, Etchegaray JI, Xie M, Zhan L, Li Y, Zhou Y, Le D, Bahr BA et al (2020) Discovery of small molecules that normalize the transcriptome and enhance cysteine cathepsin activity in progranulin-deficient microglia. Sci Rep 10:13688. https://doi.org/10.1038/s41598-020-70534-9
van der Zee J, Van Broeckhoven C (2011) TMEM106B a novel risk factor for frontotemporal lobar degeneration. J Mol Neurosci MN 45:516–521. https://doi.org/10.1007/s12031-011-9555-x
Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, Kriegstein AR (2019) Single-cell genomics identifies cell type–specific molecular changes in autism. J Sci 364:685–689. https://doi.org/10.1126/science.aav8130
Walker MP, LaFerla FM, Oddo SS, Brewer GJ (2013) Reversible epigenetic histone modifications and Bdnf expression in neurons with aging and from a mouse model of Alzheimer’s disease. Age (Dordr) 35:519–531. https://doi.org/10.1007/s11357-011-9375-5
Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031. https://doi.org/10.1016/j.cell.2009.06.049
Wauters E, Van Mossevelde S, Van der Zee J, Cruts M, Van Broeckhoven C (2017) Modifiers of GRN-associated frontotemporal lobar degeneration. Trends Mol Med 23:962–979. https://doi.org/10.1016/j.molmed.2017.08.004
Wu J, Wang SH, Potter D, Liu JC, Smith LT, Wu YZ, Huang TH, Plass C (2007) Diverse histone modifications on histone 3 lysine 9 and their relation to DNA methylation in specifying gene silencing. BMC Genomics 8:131. https://doi.org/10.1186/1471-2164-8-131
Yousef A, Robinson JL, Irwin DJ, Byrne MD, Kwong LK, Lee EB, Xu Y, Xie SX, Rennert L, Suh E et al (2017) Neuron loss and degeneration in the progression of TDP-43 in frontotemporal lobar degeneration. Acta Neuropathol Commun 5:68. https://doi.org/10.1186/s40478-017-0471-3
Zhang J, Velmeshev D, Hashimoto K, Huang YH, Hofmann JW, Shi X, Chen J, Leidal AM, Dishart JG, Cahill MK et al (2020) Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency. Nature. https://doi.org/10.1038/s41586-020-2709-7
Zhang YJ, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, O’Raw AD, Pickles SR, Prudencio M, Carlomagno Y et al (2019) Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science. https://doi.org/10.1126/science.aav2606
Zheng Y, Liu A, Wang ZJ, Cao Q, Wang W, Lin L, Ma K, Zhang F, Wei J, Matas E et al (2019) Inhibition of EHMT1/2 rescues synaptic and cognitive functions for Alzheimer’s disease. Brain 142:787–807. https://doi.org/10.1093/brain/awy354
Zhou X, Sun L, Bracko O, Choi JW, Jia Y, Nana AL, Brady OA, Hernandez JCC, Nishimura N, Seeley WW et al (2017) Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat Commun 8:15277. https://doi.org/10.1038/ncomms15277
Zhu C, Preissl S, Ren B (2020) Single-cell multimodal omics: the power of many. Nat Methods 17:11–14. https://doi.org/10.1038/s41592-019-0691-5
Acknowledgements
We acknowledge National Bioinformatics Infrastructure Sweden (NBIS, https://nbis.se/) for their support in the CITE-Seq data analysis. CITE-Seq library preparation and sequencing was performed together with the PLA and Single Cell Proteomics Facility, Uppsala University, SciLifeLab and the SNP&SEQ Technology Platform. The SNP&SEQ platform is part of the National Genomics Infrastructure (NGI) Sweden and SciLifeLab. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. WGS data was generated in NGI-SciLifeLab, Stockholm. We are grateful for support from the following funding agencies Schörling Foundation-Swedish FTD Initiative, Swedish research council (2015-02926, 2018-02754 and 2019-02248: JPND GENFI-PROX), Karolinska Institutet StratNeuro postdoc funding, Swedish Alzheimer’s foundation, Brain Foundation, Demensfonden, Karolinska geriatric foundation, Stiftelsen för Gamla Tjänarinnor, Stohnes foundation, and Region Stockholm (ALF project). We also acknowledge the patients and families, the Brain Bank at Karolinska Institutet which is supported by a CIMED-grant from Region Stockholm and associate professor Inger Nennesmo for the clinical neuropathological examinations.
Funding
Open access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Contributions
The overall study was designed by KN and CG, KN and MH conceptualized the nuclei CITE-Seq. KN performed the experiments, data interpretation and wrote first draft of the manuscript. JE did the CITE-Seq data analysis in collaboration with NBIS. Nuclei hashing experiments and antibody selection was performed by KN and MH. The generation of ADT (Antibody derived tags) was performed by MH at the PLA and Single-cell Proteomics facility at SciLifeLab, Uppsala. KP contributed with IHC analysis. JM performed ELISA of GRN, Sanger sequencing and ddPCR analysis. BK performed Western blot analysis. CG and LÖ contributed with the tissue samples and with clinical data for the study subjects. All authors contributed to the writing and read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
Nothing to report.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplementary methods and materials.
Additional file 2
. Supplementary tables.
Additional file 3
. Supplementary figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Natarajan, K., Eisfeldt, J., Hammond, M. et al. Single-cell multimodal analysis in a case with reduced penetrance of Progranulin-Frontotemporal Dementia. acta neuropathol commun 9, 132 (2021). https://doi.org/10.1186/s40478-021-01234-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40478-021-01234-2