Abstract
Background
Hereditary hearing loss is a rare hereditary condition that has a significant presence in consanguineous populations. Despite its prevalence, hearing loss is marked by substantial genetic diversity, which poses challenges for diagnosis and screening, particularly in cases with no clear family history or when the impact of the genetic variant requires functional analysis, such as in the case of missense mutations and UTR variants. The advent of next-generation sequencing (NGS) has transformed the identification of genes and variants linked to various conditions, including hearing loss. However, there remains a high proportion of undiagnosed patients, attributable to various factors, including limitations in sequencing coverage and gaps in our knowledge of the entire genome, among other factors. In this study, our objective was to comprehensively identify the spectrum of genes and variants associated with hearing loss in a cohort of 106 affected individuals from the UAE.
Results
In this study, we investigated 106 sporadic cases of hearing impairment and performed genetic analyses to identify causative mutations. Screening of the GJB2 gene in these cases revealed its involvement in 24 affected individuals, with specific mutations identified. For individuals without GJB2 mutations, whole exome sequencing (WES) was conducted. WES revealed 33 genetic variants, including 6 homozygous and 27 heterozygous DNA changes, two of which were previously implicated in hearing loss, while 25 variants were novel. We also observed multiple potential pathogenic heterozygous variants across different genes in some cases. Notably, a significant proportion of cases remained without potential pathogenic variants.
Conclusions
Our findings confirm the complex genetic landscape of hearing loss and the limitations of WES in achieving a 100% diagnostic rate, especially in conditions characterized by genetic heterogeneity. These results contribute to our understanding of the genetic basis of hearing loss and emphasize the need for further research and comprehensive genetic analyses to elucidate the underlying causes of this condition.
Similar content being viewed by others
Introduction
Hearing loss (HL) is an etiologically heterogeneous sensory deficit with genetic predisposition being a key factor in most congenital cases [1]. Although next-generation sequencing (NGS) panels and whole exome sequencing (WES) have been instrumental screening approaches during the past decade, the complexity of hearing loss genetics continues to be a diagnostic obstacle. Excluding genes involved in syndromic forms of HL, over 120 genes have been linked to non-syndromic HL so far (https://hereditaryhearingloss.org/). The reported mutational spectrum associated with these genes mainly includes point mutations, insertions and deletions (indels) as well as copy number variations (CNVs) [2, 3]. This genetic diversity often creates phenotypic variability among patients, which further complicates the diagnostic process. This is particularly evident when different mutations in the same gene lead to varying hearing loss severities, or when the same mutation results in intra-familial variability [4,5,6]. Phenotypic variability is also seen in some genes linked to both syndromic and non-syndromic HL such as MYO7A, and CDH23 [7, 8]. Additionally, while most HL genes have distinct inheritance patterns, several genes have been linked to more than one mode of inheritance. These issues collectively make the establishment of genotype-phenotype correlations often challenging [9, 10].
The use of NGS approaches for HL diagnosis is also hindered by low coverage in regions with high GC content, homology, and DNA complexity as well as limitations in our knowledge of the genome [11]. This is especially important for the detection of CNVs in genes such as STRC and OTOA which have very homologous pseudogenes. These regions of high homology greatly impact variant calling and lead to recurrent deletions and duplications as a result of non-allelic homologous recombination [2, 12, 13]. Therefore, combining NGS with additional CNV detection strategies such as multiplex ligation-dependent probe amplification (MLPA), droplet digital PCR, chromosomal microarray, and allele-specific PCR is often recommended to improve diagnostic accuracy [14].
Ensuring that a mutation completely segregates with the HL phenotype is another important aspect of making a conclusive genetic diagnosis. However, in many cases the unavailability of family history, lack of samples from important family members as well as segregation of the suspected variant with only some of the affected family members represent additional diagnostic hurdles [15]. Moreover, the emergence of cases displaying digenic inheritance where more than one pathogenic mutation appears to segregate with the HL phenotype has also been a point of discussion in recent years. The presence of digenic cases may reflect the multiple roles played by genes critical to the hearing process, as well as their ability to interact or perform co-dependent functions [16,17,18]. Nonetheless, limited knowledge of the auditory mechanism’s molecular control highlights the necessity for further functional studies. These studies can verify digenic claims and clarify the impact of mutations, specifically in the case of ambiguous missense and UTR variants.
In this study, 106 sporadic HL cases from the UAE population were screened for mutations using a combination of Sanger sequencing and WES. Though our screening approach reinforced the important contribution of GJB2 mutations among hereditary HL patients in the UAE, it also revealed relevant variants in over 20 other HL genes. Despite these findings, many cases remained undiagnosed, underscoring the intricacy of HL genetics and the shortcomings of WES when used as the sole diagnostic approach.
Materials and methods
Sample collection and GJB2 gene screening
One hundred and six sporadic cases with congenital hearing loss, primarily residing in Sharjah, Dubai, and Al Ain, were recruited from three different organizations for the Deaf and Hard of Hearing in the UAE. The recruitment period spanned from October 11, 2021, to February 23, 2023. Informed written consent was obtained from all participants and their genomic DNA was extracted from their saliva using the Oragene-DNA (OG-500) Kit (DNA Genotek, Canada). This study was approved by the Sharjah Research Ethics Committee at the University of Sharjah, Sharjah, United Arab Emirates. All affected individuals were screened for GJB2 variants using Sanger sequencing as previously described [19].
Determining cis/trans configuration of GJB2 variants
To check the cis/trans configuration of c.235delC and c.299_300delAT GJB2 variants, we cloned the PCR product of the sample showing these two variants in the heterozygous state into the pGEM-T Easy vector (Promega, USA) according to the manufacturer instructions and recombinant clones were then analyzed by Sanger sequencing.
Whole exome sequencing
Affected individuals with no GJB2 mutations were analyzed by WES using the Illumina HiSeq 2500 system as previously described [20]. In summary, exome was captured using the SureSelect All Exon V5 kit. The resulting reads passing quality control were mapped to the human reference genome, and variant calling was performed using Genome analysis tool kit (GATK) v2.7.2. Variants were annotated and filtered based on read depth and frequency in various databases. Finally, the functional impact of candidate variants was predicted using several bioinformatics tools including Variant Effect Predictor (VEP), Mutation Taster, VarSome, PROVEAN, PolyPhen-2, SIFT, and Human Splicing Finder.
Sanger sequencing.
To screen the GJB2 gene and validate variants detected by WES and recombinant plasmids, Sanger sequencing was performed. In brief, after PCR amplification, amplicons were treated with ExoSAP-IT PCR Product Cleanup Reagent (78200.200.UL, Applied Biosystems, Thermo Fisher Scientific, USA) and subsequently used in the sequencing reactions conducted using the BigDye Terminator v3.1 Cycle Sequencing Kit (4337455, Applied Biosystems, Thermo Fisher Scientific, USA). After purification, the sequencing reactions were analyzed using the Genetic Analyzer 3500 (Applied Biosystems, Thermo Fisher Scientific, USA).
Results
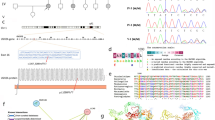
Screening of the GJB2 gene in 106 sporadic cases revealed the involvement of this gene in 24 affected individuals (Fig. 1). In fact, 20 cases were homozygous for the c.35delG (p.Gly12Valfs*2) mutation, 3 homozygous for the c.506G > A (p.Cys169Tyr) and one patient was heterozygous for the c.235delC (p.Leu79Cysfs*3) and c.299_300delAT (p.His100Argfs*14) pathogenic variants (Table 1). Given the absence of his parents’ DNA samples, we cloned the PCR product into the pGEM-T Easy vector and found after sequencing recombinant clones that c.235delC and c.299_300delAT are in trans. These results confirmed the implication of these two truncating mutations in the hearing loss phenotype observed in this affected individual.
Screening cascade used in this study. This figure illustrates the study’s screening process: All 106 hearing loss cases were initially screened for GJB2 gene mutations. Cases with detected mutations were classified accordingly. In the absence of mutations, further whole exome sequencing was conducted, and the resulting DNA variants were selectively analyzed to identify those that are rare, truncating, or classified as pathogenic or likely pathogenic within HL genes
For the rest of the affected individuals with no GJB2 mutations, we conducted whole exome sequencing (Fig. 1). Our analysis revealed the presence of 6 homozygous variants located in HL-related genes in 7 patients. Three out of these 6 variants were implicated in previous studies with HL, whereas 3 were not (Table 2). These variants are the 3 new missense variants c.3133 C > T (p.Arg1045Cys), c.934 C > T (p.Arg312Trp), c.6503T > G (p.Leu2168Arg) located in the TRIOBP, CDC14A and MYO15A genes, respectively.
Furthermore, we detected seven affected individuals with multiple heterozygous variants within HL-associated genes. Among this group, one patient (Sample # 8, as shown in Table 3) exhibited a potential segregation of two DNA variants in trans within the OTOF gene. For the remaining cases, multiple potential pathogenic heterozygous variants were observed across various genes. Moreover, in 12 patients only one heterozygous variant was detected (Table 3) with two nonsense, one frameshift and 9 missense variants. Finally, in 56 patients no potential pathogenic variant was detected in HL-associated genes.
Discussion
In this study, we investigated 106 sporadic cases affected with HL. To identify the causative mutation, Sanger sequencing of the GJB2 gene, the most common autosomal recessive non-syndromic hearing loss gene in the UAE population [21] was performed using their genomic DNA. Our analysis revealed that 20 cases were homozygous for the c.35delG mutation, 3 homozygous for the c.506G > A and one patient was compound heterozygous for the c.235delC and c.299_300delAT pathogenic variants. The c.35delG and c.506G > A have been reported previously in the UAE population [21], however this is the first time where we observe the segregation of c.235delC and c.299_300delAT in the UAE. These two mutations are the most common GJB2 mutations found in the Chinese populations [22] and were detected in neighboring populations [23, 24].
Whole exome sequencing unveiled the presence of the c.100 C > T (p.Arg34*) mutation in two cases, a variant located within the TMC1 gene. This mutation has been previously documented in numerous affected families hailing from regions such as Algeria, Tunisia, Turkey, Lebanon, Iraq, Iran, Pakistan, and Saudi Arabia [25,26,27,28,29,30]. Furthermore, it has been identified as a founder mutation in several populations [25, 29, 30].
Another confirmed mutation within our cohort was the nonsense variant c.1055 C > A (p.Ser352*), situated in the GPSM2 gene. This mutation was previously reported in a Yemeni family with Chudley-McCullough syndrome, characterized by profound congenital sensorineural hearing loss and various brain abnormalities [31].
Additionally, we identified one sample carrying the missense CDH23 mutation c.6614 C > T (p.Pro2205Leu) in the homozygous state. This variant was also noted in the homozygous state among three consanguineous probands from Qatar and in one sporadic case as a compound heterozygote in the United States [32, 33].
Moreover, our WES analysis unveiled three homozygous variants with the potential to cause disease in three cases. These changes, namely c.3133 C > T (p.Arg1045Cys), c.934 C > T (p.Arg312Trp), and c.6503T > G (p.Leu2168Arg), are located in the TRIOBP, CDC14A, and MYO15A genes, respectively. These genes are associated with three distinct forms of autosomal recessive non-syndromic hearing loss, namely DFNB28, DFNB32, and DFNB3 [34,35,36].
It is noteworthy that for the CDC14A variant (p.Arg312Trp), two mutations involving the amino acid arginine at position 312 have been previously reported: the p.Arg312Gly mutation in an Iranian family with hearing loss and confirmed infertility, and the p.Arg312Gln mutation in a Tunisian family with hearing loss, though fertility was not assessed [37]. In our study, the newly identified mutation, p.Arg312Trp, affects the same amino acid. Interestingly, the patient is a biological father, which indicates that this genetic alteration impacts hearing loss but not fertility. This may suggest that the substitution of arginine 312 with tryptophan retains some enzymatic function. This aligns with prior findings that infertility in some deaf males is linked to specific variants of CDC14A. These variants are associated with the monogenic syndrome Hearing Impairment and Infertile Male Syndrome (HIIMS), caused by inadequate phosphatase activity. In contrast, other variants with residual enzymatic function are implicated in non-syndromic deafness (DFNB32) [37].
Regarding the remaining samples, WES uncovered the presence of more than one heterozygous variant in seven cases. In sample 8, we detected two potential HL-associated variants within the OTOF gene in a heterozygous state. It’s noteworthy that compound heterozygous mutations in the OTOF gene are frequently observed and have been implicated in many cases of hearing loss [38,39,40,41,42,43,44].
Upon evaluating six samples with WES, we discovered multiple potential pathogenic heterozygous variants in genes associated with HL: In sample # 9, we identified two novel truncating duplications in a heterozygous state in two genes: USH2A c.3812-3_3837dup (p.Met1280*) and PDZD7 c.166dup (p.Arg56Profs*24). The proteins encoded by these two genes interact within the ankle region of stereocilia and play a significant role in hair cell development [45, 46]. In previous studies, a similar case was observed for the PDZD7 gene, where a heterozygous truncating mutation, p.Cys732Leufs*18, occurred alongside another heterozygous frameshift mutation, p.Ala5713Leufs*3, in the Usher protein ADGRV1 in patients with hearing loss [46]. For samples 10, 11, 12, 13, and 14, we observed multiple potential pathogenic heterozygous variants across HL-associated genes; however, no specific interactions have been reported between the implicated genes. Although numerous potential digenic interactions contributing to HL have been reported [16,17,18, 46, 48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63], confirming or refuting the role of the multigenic variants observed in patients 9, 10, 11, 12, 13 and 14 requires further segregation analyses and functional tests.
In the case of twelve samples, our WES analysis revealed the presence of only one potential heterozygous pathogenic variant. Among these 12 detected variants, three were truncating DNA changes: the well-known MYO15A nonsense variant c.5925G > A (p.Trp1975*), the novel TPRN frameshift alteration c.117del (p.Ala41Argfs*409), and the new nonsense OTOG variant c.4693G > T (p.Gly1565*). The remaining DNA variations consisted of missense variants, and none of them had been previously reported as causative for HL. It’s worth noting that while most of the genes identified in these 12 patients were associated with autosomal recessive hearing loss, mutations in the MYO7A gene have been linked to both autosomal dominant and recessive forms [9].
In our cohort, we found that the GJB2 gene accounted for 22.6% (24 out of 106) of individuals with hearing loss. This prevalence surpasses the estimates from our previous studies, where it was believed to be 18% [21]. Additionally, our analysis revealed HL-related mutations in the TMC1, GPSM2, and CDH23 genes. When combined with data from prior studies, our findings reveal that a total of 29 genes have now been confirmed to play a role in hearing loss in the UAE population [64, 65]. Notably, the most prevalent genes contributing to this condition include GJB2, COL11A2, MYO6, TMC1, TRIOBP, and TMPRSS3 [64]. Through WES, we managed to identify confirmed and potential disease-causing genotypes in approximately 25% of the individuals we examined. These findings align with previous research, highlighting the limitations of WES in identifying pathogenic mutations in all affected individuals, particularly in sporadic cases. For instance, a study by Mutai et al. in 2022 successfully identified the responsible gene in 21 out of 72 cases (approximately 29%) using WES [66]. Similarly, Reiss et al. demonstrated that WES analysis of 71 probands with hearing loss revealed pathogenic or likely pathogenic variants in only 21.1% of cases [67].
The high number of cases lacking potential pathogenic variants (~ 55%) may be attributed to the constraints of WES. These constraints include difficulties in achieving complete coverage of coding regions, often due to factors like the high G + C content in sequences, as well as limitations in detecting copy number variants. Additionally, unresolved cases, even sporadic ones, might be due to non-genetic causes of hearing loss, such as childhood infections or acoustic trauma. Other undetectable mutations besides CNVs, such as deep intronic mutations, transposable elements, and epigenetic factors, could also account for these unresolved cases. Determining whether variants are pathogenic, nonpathogenic, or of uncertain significance relies on existing data sources such as ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and may evolve as our understanding of the genome advances. Our results are similar to many previous studies where the diagnostic rate of WES was found to be less than 50%. For example the study conducted by Zeng et al. in 2022, which focused on a cohort comprising 152 familial cases with hearing loss, showed a diagnosis rate of merely 18.4%, with 28 out of the 152 cases yielding a definitive diagnosis identified using WES [68]. This result confirms the inherent limitations of WES in achieving a 100% diagnostic rate, particularly in diseases with high genetic heterogeneity like hearing loss.
Conclusion
In our investigation of 106 sporadic cases of HL, we detected mutations in the GJB2 gene, notably the prevalent c.35delG mutation. This highlights the significant role of this specific variant and GJB2 changes in general within the UAE population. Whole exome sequencing unveiled a spectrum of genetic pathogenic and likely pathogenic DNA variations. However, challenges remain, as a substantial portion of cases lack genetic explanations, underscoring the complexity of hearing loss genetics.
Data availability
All data generated or analyzed during this study can be obtained from the corresponding author on request.
Abbreviations
- HL:
-
Hearing loss
- NGS:
-
Next-generation sequencing
- WES:
-
Whole exome sequencing
- Indels:
-
Insertions and deletions
- CNVs:
-
Copy number variations
- UAE:
-
United Arab Emirates
- PCR:
-
Polymerase chain reaction
- GATK:
-
Genome analysis tool kit
- VEP:
-
Variant Effect Predictor
- SIFT:
-
Sorting Intolerant From Tolerant
- MLPA:
-
Multiplex Ligation-dependent Probe Amplification
References
Smith RJ, Bale JF Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365(9462):879–90.
Shearer AE, Kolbe DL, Azaiez H, Sloan CM, Frees KL, Weaver AE, Clark ET, Nishimura CJ, Black-Ziegelbein EA, Smith RJH. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6(5):37.
Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, Shearer AE, Sloan-Heggen CM, Kolbe D, Casavant T, Schnieders MJ, Nishimura C, Braun T, Smith RJH. Genomic Landscape and Mutational signatures of Deafness-Associated genes. Am J Hum Genet. 2018;103(4):484–97.
Back D, Shehata-Dieler W, Vona B, Hofrichter MAH, Schroeder J, Haaf T, Rahne T, Hagen R, Schraven SP. Phenotypic characterization of DFNB16-associated hearing loss. Otol Neurotol. 2019;40(1):e48–55.
Bartsch O, Vatter A, Zechner U, Kohlschmidt N, Wetzig C, Baumgart A, Nospes S, Haaf T, Keilmann A. GJB2 mutations and genotype-phenotype correlation in 335 patients from Germany with nonsyndromic sensorineural hearing loss: evidence for additional recessive mutations not detected by current methods. Audiol Neurootol. 2010;15(6):375–82.
Fujioka M, Hosoya M, Nara K, Morimoto N, Sakamoto H, Otsu M, Nakano A, Arimoto Y, Masuda S, Sugiuchi T, Masuda S, Morita N, Ogawa K, Kaga K, Matsunaga T. Differences in hearing levels between siblings with hearing loss caused by GJB2 mutations. Auris Nasus Larynx. 2020;47(6):938–42.
Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, Wayne S, Bellman S, Desmukh D, Ahmed Z, Khan SN, Kaloustian VM, Li XC, Lalwani A, Riazuddin S, Bitner-Glindzicz M, Nance WE, Liu XZ, Wistow G, Smith RJ, Griffith AJ, Wilcox ER, Friedman TB, Morell RJ. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68(1):26–37.
Weil D, Küssel P, Blanchard S, Lévy G, Levi-Acobas F, Drira M, Ayadi H, Petit C. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet. 1997;16(2):191–3.
Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet. 1997;17(3):268–9.
Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387(6628):80–3.
Butz M, McDonald A, Lundquist PA, Meyer M, Harrington S, Kester S, Stein MI, Mistry NA, Zimmerman Zuckerman E, Niu Z, Schimmenti L, Hasadsri L, Boczek NJ. Development and validation of a next-generation sequencing panel for syndromic and nonsyndromic hearing loss. J Appl Lab Med. 2020;5(3):467–79.
Laurent S, Gehrig C, Nouspikel T, Amr SS, Oza A, Murphy E, Vannier A, Béna FS, Carminho-Rodrigues MT, Blouin J, Cao Van H, Abramowicz M, Paoloni-Giacobino A, Guipponi M. Molecular characterization of pathogenic OTOA gene conversions in hearing loss patients. Hum Mutat. 2021;42(4):373–7.
Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81.
Shi L, Bai Y, Kharbutli Y, Oza AM, Amr SS, Edelmann L, Mehta L, Scott SA. Prenatal cytogenomic identification and molecular refinement of compound heterozygous STRC deletion breakpoints. Mol Genet Genomic Med. 2019;7(8):e806.
Zonta F, Girotto G, Buratto D, Crispino G, Morgan A, Abdulhadi K, Alkowari M, Badii R, Gasparini P, Mammano F. The p.Cys169Tyr variant of connexin 26 is not a polymorphism. Hum Mol Genet. 2015;24(9):2641–8.
Schrauwen I, Chakchouk I, Acharya A, Liaqat K, Irfanullah, University of Washington Center for Mendelian Genomics, Nickerson DA, Bamshad MJ, Shah K, Ahmad W, Leal SM. Novel digenic inheritance of PCDH15 and USH1G underlies profound non-syndromic hearing impairment. BMC Med Genet. 2018;19(1):122–5.
Leone MP, Palumbo P, Ortore R, Castellana S, Palumbo O, Melchionda S, Palladino T, Stallone R, Mazza T, Cocchi R, Carella M. Putative TMPRSS3/GJB2 digenic inheritance of hearing loss detected by targeted resequencing. Mol Cell Probes 2017, 3324–27.
Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, Chang B, Johnson KR, Liu XZ. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet. 2005;14(1):103–11.
Mahfood M, Chouchen J, Kamal EA, Mohamed, Walaa, Al Mutery A, Harati R, Tlili A. Whole exome sequencing, in silico and functional studies confirm the association of the GJB2 mutation p.Cys169Tyr with deafness and suggest a role for the TMEM59 gene in the hearing process. Saudi J Biol Sci. 2021;28(8):4421–9.
Mahfood M, Chouchen J, Kamal Eddine Ahmad Mohamed W, Al Mutery A, Harati R, Tlili A. Whole exome sequencing, in silico and functional studies confirm the association of the GJB2 mutation p.Cys169Tyr with deafness and suggest a role for the TMEM59 gene in the hearing process. Saudi J Biol Sci. 2021;28(8):4421–9.
Tlili A, Al Mutery A, Kamal Eddine Ahmad Mohamed W, Mahfood M, Hadj Kacem H. Prevalence of GJB2 mutations in affected individuals from United Arab Emirates with autosomal recessive nonsyndromic hearing loss. Genetic Test Mol Biomarkers. 2017;21(11):686–91.
Zhang Y, Wang J, Li L, Sun Y, Feng B. Three common GJB2 mutations causing nonsyndromic hearing loss in Chinese populations are retained in the endoplasmic reticulum. Acta Otolaryngol. 2010;130(7):799–803.
Azadegan-Dehkordi F, Bahrami T, Shirzad M, Karbasi G, Yazdanpanahi N, Farrokhi E, Koohiyan M, Tabatabaiefar MA, Hashemzadeh-Chaleshtori M. Mutations in GJB2 as major causes of autosomal recessive non-syndromic hearing loss: First Report of c.299-300delAT mutation in kurdish Population of Iran. J Audiol Otol. 2019;23(1):20–6.
Koohiyan M, Azadegan-Dehkordi F, Koohian F, Hashemzadeh-Chaleshtori M. Genetics of hearing loss in North Iran Population: an update of spectrum and frequency of GJB2 mutations. J Audiol Otol. 2019;23(4):175–80.
Kitajiri SI, McNamara R, Makishima T, Husnain T, Zafar AU, Kittles RA, Ahmed ZM, Friedman TB, Riazuddin S, Griffith AJ. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet. 2007;72(6):546–50.
Hilgert N, Alasti F, Dieltjens N, Pawlik B, Wollnik B, Uyguner O, Delmaghani S, Weil D, Petit C, Danis E, Yang T, Pandelia E, Petersen MB, Goossens D, Favero JD, Sanati MH, Smith RJ, Van Camp G. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet. 2008;74(3):223–32.
Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, Ghosh M, Menon PS, Deshmukh D, Oddoux C, Ostrer H, Khan S, Riazuddin S, Deininger PL, Hampton LL, Sullivan SL, Battey JF Jr, Keats BJ, Wilcox ER, Friedman TB, Griffith AJ. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30(3):277–84.
Sirmaci A, Duman D, Ozturkmen-Akay H, Erbek S, Incesulu A, Ozturk-Hismi B, Arici ZS, Yuksel-Konuk EB, Tasir-Yilmaz S, Tokgoz-Yilmaz S, Cengiz FB, Aslan I, Yildirim M, Hasanefendioglu-Bayrak A, Aycicek A, Yilmaz I, Fitoz S, Altin F, Ozdag H, Tekin M. Mutations in TMC1 contribute significantly to nonsyndromic autosomal recessive sensorineural hearing loss: a report of five novel mutations. Int J Pediatr Otorhinolaryngol. 2009;73(5):699–705.
Tlili A, Rebeh IB, Aifa-Hmani M, Dhouib H, Moalla J, Tlili-Chouchene J, Said MB, Lahmar I, Benzina Z, Charfedine I, Driss N, Ghorbel A, Ayadi H, Masmoudi S. TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol Neurootol. 2008;13(4):213–8.
Ramzan K, Al-Owain M, Al-Numair NS, Afzal S, Al-Ageel S, Al-Amer S, Al-Baik L, Al-Otaibi GF, Hashem A, Al-Mashharawi E, Basit S, Al-Mazroea AH, Softah A, Sogaty S, Imtiaz F. Identification of TMC1 as a relatively common cause for nonsyndromic hearing loss in the Saudi population. Am J Med Genet B Neuropsychiatr Genet. 2020;183(3):172–80.
Hamzeh AR, Nair P, Mohamed M, Saif F, Tawfiq N, Al-Ali MT, Bastaki F. A novel nonsense GPSM2 mutation in a Yemeni family underlying Chudley-McCullough syndrome. Eur J Med Genet. 2016;59(6–7):337–41.
Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, Ephraim SS, Shibata SB, Booth KT, Campbell CA, Ranum PT, Weaver AE, Black-Ziegelbein EA, Wang D, Azaiez H, Smith RJH. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135(4):441–50.
Alkowari MK, Vozzi D, Bhagat S, Krishnamoorthy N, Morgan A, Hayder Y, Logendra B, Najjar N, Gandin I, Gasparini P, Badii R, Girotto G, Abdulhadi K. Targeted sequencing identifies novel variants involved in autosomal recessive hereditary hearing loss in Qatari families. Mutat Res 2017, 800–2;29–36.
Delmaghani S, Aghaie A, Bouyacoub Y, El Hachmi H, Bonnet C, Riahi Z, Chardenoux S, Perfettini I, Hardelin J, Houmeida A, Herbomel P, Petit C. Mutations in CDC14A, encoding a protein phosphatase involved in hair cell ciliogenesis, cause autosomal-recessive severe to Profound Deafness. Am J Hum Genet. 2016;98(6):1266–70.
Shahin H, Walsh T, Sobe T, Abu Sa’ed J, Abu Rayan A, Lynch ED, Lee MK, Avraham KB, King M, Kanaan M. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am J Hum Genet. 2006;78(1):144–52.
Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280(5368):1447–51.
Imtiaz A, Belyantseva IA, Beirl AJ, Fenollar-Ferrer C, Bashir R, Bukhari I, Bouzid A, Shaukat U, Azaiez H, Booth KT, Kahrizi K, Najmabadi H, Maqsood A, Wilson EA, Fitzgerald TS, Tlili A, Olszewski R, Lund M, Chaudhry T, Rehman AU, Starost MF, Waryah AM, Hoa M, Dong L, Morell RJ, Smith RJH, Riazuddin S, Masmoudi S, Kindt KS, Naz S, Friedman TB. CDC14A phosphatase is essential for hearing and male fertility in mouse and human. Hum Mol Genet. 2018;27(5):780–98.
Qiu Y, Chen S, Xie L, Xu K, Lin Y, Bai X, Zhang H, Liu X, Jin Y, Sun Y, Kong W. Auditory Neuropathy Spectrum Disorder due to two novel compound heterozygous OTOF mutations in two Chinese families. Neural Plast. 2019;2019:9765276.
Iwasa Y, Nishio S, Sugaya A, Kataoka Y, Kanda Y, Taniguchi M, Nagai K, Naito Y, Ikezono T, Horie R, Sakurai Y, Matsuoka R, Takeda H, Abe S, Kihara C, Ishino T, Morita S, Iwasaki S, Takahashi M, Ito T, Arai Y, Usami S. OTOF mutation analysis with massively parallel DNA sequencing in 2,265 Japanese sensorineural hearing loss patients. PLoS ONE. 2019;14(5):e0215932.
Bai X, Nian S, Feng L, Ruan Q, Luo X, Wu M, Yan Z. Identification of novel variants in MYO15A, OTOF, and RDX with hearing loss by next-generation sequencing. Mol Genet Genomic Med. 2019;7(8):e808.
Ren S, Kong X, Shi H. Application of next generation sequencing and Sanger sequencing in a pedigree affected with hereditary non-syndromic deafness. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2018;35(6):864–7.
Wang Y, Lu Y, Cheng J, Zhang L, Han D, Yuan H. Novel OTOF gene mutations identified using a massively parallel DNA sequencing technique in DFNB9 deafness. Acta Otolaryngol. 2018;138(10):865–70.
Tang F, Ma D, Wang Y, Qiu Y, Liu F, Wang Q, Lu Q, Shi M, Xu L, Liu M, Liang J. Novel compound heterozygous mutations in the OTOF Gene identified by whole-exome sequencing in auditory neuropathy spectrum disorder. BMC Med Genet 2017, 18(1);35 – 0.
Zhang L, Chai Y, Yang T, Wu H. Identification of novel OTOF compound heterozygous mutations by targeted next-generation sequencing in a Chinese patient with auditory neuropathy spectrum disorder. Int J Pediatr Otorhinolaryngol. 2013;77(10):1749–52.
Zou J, Zheng T, Ren C, Askew C, Liu X, Pan B, Holt JR, Wang Y, Yang J. Deletion of PDZD7 disrupts the Usher syndrome type 2 protein complex in cochlear hair cells and causes hearing loss in mice. Hum Mol Genet. 2014;23(9):2374–90.
Ebermann I, Phillips JB, Liebau MC, Koenekoop RK, Schermer B, Lopez I, Schäfer E, Roux A, Dafinger C, Bernd A, Zrenner E, Claustres M, Blanco B, Nürnberg G, Nürnberg P, Ruland R, Westerfield M, Benzing T, Bolz HJ. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J Clin Invest. 2010;120(6):1812–23.
Lerer I, Sagi M, Ben-Neriah Z, Wang T, Levi H, Abeliovich D. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: a novel founder mutation in Ashkenazi jews. Hum Mutat. 2001;18(5):460.
Gürtler N, Egenter C, Bösch N, Plasilova M. Mutation analysis of the Cx26, Cx30, and Cx31 genes in autosomal recessive nonsyndromic hearing impairment. Acta Otolaryngol. 2008;128(10):1056–62.
Liu X, Yuan Y, Yan D, Ding EH, Ouyang XM, Fei Y, Tang W, Yuan H, Chang Q, Du LL, Zhang X, Wang G, Ahmad S, Kang DY, Lin X, Dai P. Digenic inheritance of non-syndromic deafness caused by mutations at the gap junction proteins Cx26 and Cx31. Hum Genet. 2009;125(1):53–62.
Cama E, Melchionda S, Palladino T, Carella M, Santarelli R, Genovese E, Benettazzo F, Zelante L, Arslan E. Hearing loss features in GJB2 biallelic mutations and GJB2/GJB6 digenic inheritance in a large Italian cohort. Int J Audiol. 2009;48(1):12–7.
Kooshavar D, Tabatabaiefar MA, Farrokhi E, Abolhasani M, Noori-Daloii M, Hashemzadeh-Chaleshtori M. Digenic inheritance in autosomal recessive non-syndromic hearing loss cases carrying GJB2 heterozygote mutations: assessment of GJB4, GJA1, and GJC3. Int J Pediatr Otorhinolaryngol. 2013;77(2):189–93.
Lechowicz U, Pollak A, Oziębło D, Ołdak M. Pathogenic p.Cys194Metfs*17 variant argues against TMPRSS3/GJB2 digenic inheritance of hearing loss. Eur Arch Otorhinolaryngol. 2016;273(5):1327–8.
Vona B, Lechno S, Hofrichter MAH, Hopf S, Läig AK, Haaf T, Keilmann A, Zechner U, Bartsch O. Confirmation of PDZD7 as a nonsyndromic hearing loss gene. Ear Hear. 2016;37(4):238.
Wang H, Feng Y, Li H, Wu H, Mei L, Wang X, Jiang L, He C. Digenic mutations involving both the BSND and GJB2 genes detected in Bartter syndrome type IV. Int J Pediatr Otorhinolaryngol. 2017;92:17–20.
Mei L, Chen J, Zong L, Zhu Y, Liang C, Jones RO, Zhao H. A deafness mechanism of digenic Cx26 (GJB2) and Cx30 (GJB6) mutations: reduction of endocochlear potential by impairment of heterogeneous gap junctional function in the cochlear lateral wall. Neurobiol Dis 2017, 108195–203.
Nagara M, Papagregoriou G, Ben Abdallah R, Landoulsi Z, Bouyacoub Y, Elouej S, Kefi R, Pippucci T, Voskarides K, Bashamboo A, McElreavey K, Hachicha M, Romeo G, Seri M, Deltas C, Abdelhak S. Distal renal tubular acidosis in a Libyan patient: evidence for digenic inheritance. Eur J Med Genet. 2018;61(1):1–7.
Naseri M, Akbarzadehlaleh M, Masoudi M, Ahangari N, Poursadegh Zonouzi AA, Poursadegh Zonouzi A, Shams L, Nejatizadeh A. Genetic linkage analysis of DFNB4, DFNB28, DFNB93 loci in autosomal recessive non-syndromic hearing loss: evidence for digenic inheritance in GJB2 and GJB3 mutations. Iran J Public Health. 2018;47(1):95–102.
Chen K, Wu X, Zong L, Jiang H. GJB3/GJB6 screening in GJB2 carriers with idiopathic hearing loss: is it necessary? J Clin Lab Anal. 2018;32(9):e22592.
Smits JJ, Oostrik J, Beynon AJ, Kant SG, de Koning Gans, Pia AM, Rotteveel LJC, Klein Wassink-Ruiter JS, Free RH, Maas SM, van de Kamp J, Merkus P, Consortium DOOFNL, Koole W, Feenstra I, Admiraal RJC, Lanting CP, Schraders M, Yntema HG, Pennings RJE, Kremer H. De novo and inherited loss-of-function variants of ATP2B2 are associated with rapidly progressive hearing impairment. Hum Genet. 2019;138(1):61–72.
Li M, Nishio S, Naruse C, Riddell M, Sapski S, Katsuno T, Hikita T, Mizapourshafiyi F, Smith FM, Cooper LT, Lee MG, Asano M, Boettger T, Krueger M, Wietelmann A, Graumann J, Day BW, Boyd AW, Offermanns S, Kitajiri S, Usami S, Nakayama M. Digenic inheritance of mutations in EPHA2 and SLC26A4 in Pendred syndrome. Nat Commun. 2020;11(1):1343–9.
Furlano M, Martínez V, Pybus M, Arce Y, Crespí J, Venegas MDP, Bullich G, Domingo A, Ayasreh N, Benito S, Lorente L, Ruíz P, Gonzalez VL, Arlandis R, Cabello E, Torres F, Guirado L, Ars E, Torra R. Clinical and genetic features of autosomal Dominant Alport Syndrome: a Cohort Study. Am J Kidney Dis. 2021;78(4):560–e5701.
Zhang Q, Qin T, Hu W, Janjua MU, Jin P. Putative digenic GJB2/MYO7A inheritance of hearing loss detected in a patient with 48,XXYY Klinefelter Syndrome. Hum Hered. 2020;85(3–6):117–24.
Liu M, Liang Y, Huang B, Sun J, Chen K. Report of rare and novel mutations in candidate genes in a cohort of hearing-impaired patients. Mol Genet Genomic Med. 2022;10(4):e1887.
Elsayed O. A Al-Shamsi 2022 Mutation spectrum of non-syndromic hearing loss in the UAE, a retrospective cohort study and literature review. Mol Genet Genomic Med 10 11 e2052.
Al Mutery A, Mahfood M, Chouchen J, Tlili A. Genetic etiology of hereditary hearing loss in the Gulf Cooperation Council countries. Hum Genet. 2022;141(3–4):595–605.
Mutai H, Momozawa Y, Kamatani Y, Nakano A, Sakamoto H, Takiguchi T, Nara K, Kubo M, Matsunaga T. Whole exome analysis of patients in Japan with hearing loss reveals high heterogeneity among responsible and novel candidate genes. Orphanet J Rare Dis. 2022;17(1):114–4.
Reis CS, Quental S, Fernandes S, Castedo S, Moura CP. Whole-exome sequencing targeting a Gene Panel for Sensorineural hearing loss: the First Portuguese Cohort Study. Cytogenet Genome Res. 2022;162(1–2):1–9.
Zeng B, Xu H, Yu Y, Li S, Tian Y, Li T, Yang Z, Wang H, Wang G, Chang M, Tang W. Increased diagnostic yield in a cohort of hearing loss families using a comprehensive stepwise strategy of molecular testing. Front Genet 2022, 131057293.
Acknowledgements
We would like to thank all participants for their cooperation.
Funding
This study was funded by the University of Sharjah.
Author information
Authors and Affiliations
Contributions
AT: study design, supervision, and data analysis. AT, MM, AM and JC: clinical samples and data acquisition. AT, MM and JC: experimental work, data analysis and original draft writing. AT, MM, AM and JC: manuscript revision and editing. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Sharjah Research Ethics Committee at the University of Sharjah, Sharjah, United Arab Emirates. Informed written consent was obtained from all family members or their parents if under 18 years of age.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tlili, A., Mahfood, M., Al Mutery, A. et al. Genetic analysis of 106 sporadic cases with hearing loss in the UAE population. Hum Genomics 18, 59 (2024). https://doi.org/10.1186/s40246-024-00630-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40246-024-00630-8